Abstract
Twenty inter-simple sequence repeat (ISSR) and twenty two start codon targeted (SCoT) primers were employed to analyze genetic diversity and population structure among 52 Trichosanthes dioica Roxb. accessions collected from nine different eco-geographical regions of India. ISSR markers proved to be more informative in genetic diversity assessment and produced higher mean number of polymorphic bands (15.25 with 95.96% polymorphism) and polymorphic information content (PIC) value (0.47) compared to SCoT markers (12.55 polymorphic bands with 92.20% polymorphism and PIC: 0.45). Total genetic diversity (Ht) and genetic diversity within populations (Hs) in T. dioica accessions was found to be very high (0.45 and 0.43, respectively). AMOVA analysis also revealed higher genetic variation within populations (81%) than among them (19%). Among different T. dioica populations, very low genetic differentiation (Gst: 0.05) and high gene flow (Nm: 9.32) were observed. T. dioica populations of Bihar state were found to be highly diverse and Kolkata and Cuttack populations were least diverse. T. dioica male plants were more variable than females. UPGMA, Neighbor-Joining and population structure analyses divided T. dioica populations into three main clusters. First cluster comprised of Meerut population, second cluster included of Cuttack and Kolkata populations and populations of Bihar, Delhi and Kanpur occurred in third cluster. Genetic diversity was found to be strongly positively correlated with the latitude and strongly negatively correlated with annual mean rainfall of different T. dioica cultivated regions. For sex identification, one SRAP primer combination, 'Em-6/Me-4' amplified two molecular markers of around 230 and 290 bp specific to male T. dioica plants of Bihar, Kanpur, North Delhi and Meerut populations and were completely absent from female plants.
Keywords: Genetics, Molecular biology, Ecology
1. Introduction
Trichosanthes dioica Roxb. (Pointed gourd), a member of family Cucurbitaceae is a perennial, dioecious climber herb, native to Assam-Bengal regions of India [1]. This plant is extensively cultivated for its nutritious fruits in tropical and subtropical regions of the world and in India, it is propagated all over the warmer regions particularly in Uttar Pradesh, Bihar, West Bengal and Assam [2, 3, 4]. Its fruits are used as vegetable and are rich source of proteins, vitamin A and minerals [5], easily digestible, diuretic in nature, having anti-ulcerous and anti-diabetic effects [6]. In T. dioica, cultivation through seeds is uneconomical for farmers due to poor germination, undesirable male-female sex ratio and inability to identify sex prior flowering, which takes around 7–8 months [7]. Seeds raised populations produce more male plants than females [6]. To overcome these seeds related issues, this crop is mainly propagated through vine cuttings and root suckers in the desire of obtaining more fruits in short time. However, vegetative propagation may result in loss of genetic diversity due to less occurrence of evolution and adaptation, which arise frequently in sexual reproduction. Sex identification at early seedling stages will encourage farmers to propagate T. dioica plants employing seeds, which have positive effects on genetic diversity and will save undesirable waste of time, space, resources and nursery costs of farmers.
Genetic diversity is a basic component of biodiversity [8] and its conservation is essential for long term survival of any species in changing environments. Among different populations, genetic diversity is non randomly distributed and is affected by various factors such as geographic variations, breeding systems, dispersal mechanisms, life span, etc. [9]. Change in environmental conditions often leads to variation in genetic diversity levels among different populations [10] and populations with low variability are generally considered less adapted under adverse circumstances. Therefore, genetic diversity assessment in geographically isolated populations is very valuable for identification of low diverse populations and maintaining high genetic diversity in such populations through introduction of highly variable cultivars, breeding programs, etc. Studies pertaining to genetic structure of different populations are useful for understanding historical events, population dynamics, etc. In dioecious plants, male and female reproductive organs occur on separate plants and levels of genetic variations between sexes are different [11, 12], caused by several factors such as distribution patterns of individuals across the populations [13], sex ratio [12], stochastic events [14], etc. Differences in genetic diversity levels between plant sexes can be used for making breeding programs to obtain better hybrid varieties with maximum genetic polymorphism.
Molecular markers play a significant role in protection of biodiversity, identification of promising cultivars, quantitative trait loci (QTL) mapping, etc. [15]. Different PCR based dominant markers such as ISSR, SCoT, SRAP, etc. have been effectively used for quantification of genetic diversity [16, 17, 18, 19] and sex identification in different plants [20, 21]. In the present study, ISSR and SCoT markers were employed to analyze genetic diversity and phylogenetic relationships in 52 T. dioica accessions belonging to nine different populations of India and for sex identification, SRAP markers were used.
2. Materials and methods
2.1. Plant material
For genetic diversity study, fresh leafy twigs of 52 physiologically matured T. dioica accessions belonging to nine different populations with different eco-geographic characteristics were sampled and stored in -20° till further use. More information about geographical distribution of accessions are in Table 1 and Fig. S1. All the fifty two accessions are being maintained at Delhi University herbarium, India. For sex linked diversity analysis, three male and three female plants were procured separately from populations of Bihar, Kanpur, North Delhi and Meerut.
Table 1.
Different populations and plant accessions of T. dioica used in the study.
| S. No. | Population (Collection site) | No. of collected accessions | Accession number | Geographical coordinates | Altitude (meters) | Mean maximum temperature (°C) | Mean minimum temperature (°C) |
Annual rainfall (mm) | Days to Flowering (range) | Fruit weight (g) (range) |
Fruit length (cm) (range) |
Mode of Cultivation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Kanpur (Uttar Pradesh) | 6 | DUH14128, DUH14130 (Kanpur Dehat); DUH14144, DUH14145 (Kanpur Nagar); DUH14129, DUH14142 (Farrukhabad) |
26.4499° N, 80.3319° E | 126 | 32.2 | 19.5 | 821 | 60–65 | 22–26 | 6.5–7.6 | Aerial support and flat bed |
| 2 | Samastipur (Bihar) | 6 | DUH14133, DUH14134, DUH14293, DUH14294, DUH14295, DUH14296 | 25.7471° N, 85.8896° E | 39 | 31.40 | 16.80 | 1142 | 58–60 | 32–35 | 8.1–8.8 | Aerial support |
| 3 | Saran (Bihar) | 6 | DUH14138, DUH14139, DUH14283, DUH14284, DUH14285, DUH14286 | 25.8560° N, 84.8568° E | 36 | 30.00 | 18.00 | 1059 | 57–60 | 31–35 | 8.2–8.7 | Aerial support |
| 4 | Muzaffarpur (Bihar) | 6 | DUH14135, DUH14136, DUH14281, DUH 14287, DUH 14288, DUH 14289 | 26.1209° N, 85.3647° E | 60 | 32.30 | 19.30 | 1182 | 58–62 | 32–35 | 8.1–8.7 | Aerial support |
| 5 | Gopalganj (Bihar) | 6 | DUH14137, DUH14140, DUH14282, DUH14290, DUH14291, DUH14292 | 26.4514° N, 84.3963° E | 60 | 31.50 | 16.20 | 1218 | 55–60 | 33–36 | 8.2–8.8 | Aerial support |
| 6 | North Delhi (Delhi) | 6 | DUH14127, DUH14141, DUH14297, DUH14298, DUH14299, DUH14300 | 28.7041° N, 77.1025° E | 213 | 33.00 | 13.00 | 767 | 61–65 | 25–28 | 6.9–7.8 | Flat bed |
| 7 | Meerut (Uttar Pradesh) | 12 | DUH14126, DUH14131, DUH14301, DUH14302, DUH14303, DUH14304 (Meerut); DUH14132, DUH14143, DUH14305, DUH14306, DUH14307, DUH14308 (Baghpat) | 28.9845° N, 77.7064° E | 225 | 33.50 | 14.20 | 768 | 60–66 | 24–28 | 6.7–7.5 | Flat bed |
| 8 | Kolkata (West Bengal) | 2 | DUH14309, DUH14310 | 22.5726° N, 88.3639° E | 9 | 31.80 | 22.10 | 1641 | 62–68 | 32–37 | 7.4–8.0 | Aerial support and flat bed |
| 9 | Cuttack (Odisha) | 2 | DUH14311, DUH14312 | 20.4625° N, 85.8830° E | 36 | 32.8 | 22.25 | 1800 | 65–72 | 30–36 | 7.2–8.1 | Aerial support and flat bed |
For generation of sex linked molecular marker, a total of 42 T. dioica plants (21 females and 21 males) were procured from Samastipur, Saran, Muzaffarpur, Gopalganj, Kanpur, North Delhi and Meerut populations. From each region, 3 male and 3 female plants were collected.
2.2. Genomic DNA extraction
Genomic DNA of each T. dioica accession was extracted according to modified CTAB (Cetyl Trimethyl Ammonium Bromide) method [22]. The quantity of RNase-treated DNA of all the plant accessions was determined on agarose gel (0.8%), comparing with a standard lambda DNA marker (Amersham Biosciences, Piscataway, NJ, USA).
2.3. For genetic diversity study
2.3.1. Morphological analysis
Different morphological traits such as days to flowering, fruit weight, fruit length and cultivation style were recorded (Table 1). Fifty fruits from each population were analyzed for calculating range of weight and length of the T. dioica fruits. Thirty T. dioica plants from each population were kept under observation for estimating days to flowering.
2.3.2. ISSR and SCoT-PCR amplification
Twenty ISSR and twenty two SCoT primers (Integrated DNA Technologies, Inc, USA) were randomly selected and tested on isolated genomic DNA of T. dioica accessions (Tables 2 and 3). The PCR reactions were carried out in a 25 μL volume containing Taq polymerase (1 unit), genomic DNA (50 ng), ISSR/SCoT primers (0.80 μM), dNTP (0.1 mM), 10X PCR reaction buffer (2.5 μL), Triton X (1%) and MgCl2 (3 mM). PCR amplifications were carried out with a preliminary cycle of 120 s at 94 °C, followed by 35 cycles of 30 s at 94 °C, 60 s at 50 °C, 120 s at 72 °C, and a final extension of 7 min at 72 °C. The amplification products were resolved on 1.2 % agarose gels.
Table 2.
Different ISSR primers used for detecting polymorphism among 52 T. dioica accessions.
| S. No. | Primer name | Primer sequence (5′-3′) | Total number of bands | Number of polymorphic bands | Percentage polymorphism | PIC | EMR | MI | RP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | UBC807 | AGAGAGAGAGAGAGAGT | 16 | 16 | 100% | 0.448 | 16 | 7.17 | 21.19 |
| 2 | UBC835 | AGAGAGAGAGAGAGAGYC | 14 | 12 | 85.71% | 0.432 | 10.28 | 4.44 | 18.96 |
| 3 | UBC834 | AGAGAGAGAGAGAGAGYT | 16 | 15 | 93.75% | 0.452 | 14.06 | 6.36 | 21.23 |
| 4 | UBC868 | GAAGAAGAAGAAGAAGAA | 17 | 16 | 94.12% | 0.495 | 15.06 | 7.45 | 16.92 |
| 5 | VIS11 | CACCACCACGC | 18 | 18 | 100% | 0.495 | 18 | 8.91 | 19.73 |
| 6 | VIS14 | GAGAGAGAGAGACC | 17 | 17 | 100% | 0.487 | 17 | 8.28 | 14.88 |
| 7 | VIS20 | GGAGAGGAGAGGAGA | 12 | 12 | 100% | 0.496 | 12 | 5.95 | 11.08 |
| 8 | VIS22 | VHVGTGTGTGTGTGTGT | 17 | 16 | 94.12% | 0.480 | 15.06 | 7.22 | 20.38 |
| Total | 127 | 122 | |||||||
| Mean | 15.87 | 15.25 | 95.96% | 0.47 | 14.68 | 6.97 | 18.05 |
PIC: polymorphism information content, EMR: effective multiplex ratio, MI: marker index, RP: resolving power.
Table 3.
Different SCoT primers used for detecting polymorphism among 52 T. dioica accessions.
| S. No. | Primer name | Primer sequence (5′-3′) | Total number of bands | Number of polymorphic bands | Percentage polymorphism | PIC | EMR | MI | RP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SCoT1 | CAACATGGCTACCACCA | 11 | 11 | 100% | 0.492 | 11 | 5.41 | 9.65 |
| 2 | SCoT5 | CAACAATGGCTACCACGA | 13 | 10 | 76.92% | 0.424 | 7.69 | 3.26 | 17.88 |
| 3 | SCoT13 | ACGACATGGCGACCATCG | 13 | 13 | 100% | 0.396 | 13 | 5.15 | 19.08 |
| 4 | SCoT15 | ACGACATGGCGACCGCGA | 13 | 13 | 100% | 0.487 | 13 | 6.33 | 15.42 |
| 5 | SCoT19 | ACCATGGCTACCACCGGC | 15 | 12 | 80% | 0.438 | 9.6 | 4.20 | 20.54 |
| 6 | SCoT28 | CCATGGCTACCACCGCCA | 16 | 13 | 81.25% | 0.424 | 10.56 | 4.48 | 22 |
| 7 | SCoT34 | ACCATGGCTACCACCGCA | 13 | 13 | 100% | 0.496 | 13 | 6.45 | 14.04 |
| 8 | SCoT39 | ACGACATGGCGACCAGCG | 12 | 11 | 91.67% | 0.430 | 10.08 | 4.33 | 16.65 |
| 9 | SCoT40 | ACGACATGGCGACCACGT | 17 | 17 | 100% | 0.495 | 17 | 8.41 | 18.77 |
| Total | 123 | 113 | |||||||
| Mean | 13.67 | 12.55 | 92.20% | 0.45 | 11.66 | 5.33 | 17.11 |
PIC: polymorphism information content, EMR: effective multiplex ratio, MI: marker index, RP: resolving power.
2.3.3. Data scoring and statistical analysis
Out of the twenty ISSR and twenty two SCoT primers tested, eight ISSR and nine SCoT primers produced clear and polymorphic bands and were chosen for further analysis. Banding patterns produced by eight ISSR and nine SCoT markers were scored for absence (0) and presence (1) of bands. Initially, by observing the banding patterns produced by different ISSR and SCoT primers, total number of bands, polymorphic bands and percentage polymorphism were obtained. Further, potential of these molecular markers for estimation of genetic variability was assessed by measuring polymorphism information content (PIC), effective multiplex ratio (EMR), marker index (MI) and resolving power (RP). PIC values were calculated using the formula PIC = 1- ∑pi2, where pi is the frequency of the ith allele [23]. Marker index (MI) is the primer capacity to detect polymorphic loci among different genotypes and was calculated as EMR X PIC, where, EMR is the product of number of polymorphic loci and fraction of polymorphic loci. Resolving power (RP) is the ability of primers to distinguish between genotypes and was calculated as RP = ΣIb, where Ib is the informative fragments.
Genetic diversity was measured by different parameters such as effective number of alleles (Ne), Nei's gene diversity (H), Shannon's information index (I), total genetic diversity (Ht), genetic diversity within populations (Hs), genetic differentiation (Gst) and gene flow (Nm) which were calculated using POPGENE (version 1.31) program [24]. This software was also used to construct distance/similarity matrix and Nei's genetic distance based Unweighted pair group method with arithmetic average (UPGMA) tree. UPGMA and neighbor-joining (NJ) trees comprising of fifty two T. dioica accessions were also generated by combined data of ISSR and SCoT markers employing Free Tree software [25] based on Jaccard's coefficient of similarity. Both the trees were visualized using Tree View software [26]. For analyzing population genetic structure of T. dioica accessions in India employing combined data of ISSR and SCoT markers, STRUCTURE software version 2.3.4, based on Bayesian model-based clustering [27] was used. Following parameter setting was done; Markov Chain Monte Carlo replication (MCMC Reps): 10,000; length of burnin period: 10,000; models: admixture model and correlated allele frequency. A total of 20 independent runs were performed for each value of K (from 1 to 7) [28]. Structure Harvester v6.0 program developed by Earl Dent and vonHoldt Bridgett [29] was employed to determine the best value of K for the data. Based on the best K value obtained, the number of subpopulations was determined. Analysis of molecular variance (AMOVA) was conducted on within and among populations, employing GenAlEx 6.5 software based on 999 permutations [30].
The correlations of genetic diversity parameters (Ne, H and I) with geographical coordinates (latitude, longitude and altitude) and different climatic factors (mean minimum temperature, mean maximum temperature and annual rainfall) were evaluated using SPSS version 21.0 software. The same software was used to analyze significant differences using Duncan's multiple range tests (P < 0.05) between the mean values of male and female genetic diversity parameters (Ne, H and I) and the means of ISSR and SCoT generated Ht, Hs, Gst and Nm values. All data in the present study is represented as mean ± standard deviation of the mean (SD).
2.4. For identification of sex linked molecular markers
2.4.1. Bulked segregant analysis-SRAP analysis
Initially, two bulk DNA samples (male and female separately) were prepared by pooling an equal concentration of DNA from individual plants of Bihar, Kanpur, North Delhi and Meerut regions. A total of 50 combinations of forward and reverse primers were used for BSA-SRAP analysis [31]. PCR was performed under following conditions: initial DNA denaturation (95 °C for 4 min), followed by 5 cycles of denaturation (94 °C for 1 min), annealing (36 °C for 1 min) and extension (72 °C for 2 min). For next 30 cycles, annealing temperature was increased to 50 °C with final extension of 72 °C for 7 min. Two percent agarose gels were used to resolve the PCR amplified products. A DNA marker present in either male or female bulk sample was considered to be a putative sex-linked marker.
2.4.2. Validation of the putative sex linked SRAP markers on larger populations
Putative sex linked markers were further tested on individual plants (3 male and 3 female) of Bihar (Samastipur, Saran, Muzaffarpur and Gopalganj), Kanpur, North Delhi and Meerut regions.
3. Results
3.1. For genetic diversity study
3.1.1. Morphological analysis
T. dioica accessions collected from Samastipur, Saran, Muzaffarpur and Gopalganj regions of Bihar state were being cultivated on trellis systems for aerial support (Table 1). Flat bed systems were being used for cultivation of T. dioica populations of North Delhi and Meerut regions. However, populations of Kanpur, Kolkata and Cuttack were using both the systems (trellis and flat bed) for T. dioica cultivation (Table 1). T. dioica populations of Bihar state took lesser days to produce flowers (57–62 days) compared to other populations. Fruit length and fruit weight of T. dioica populations of Bihar state were also found higher in comparison to other populations (Table 1).
3.1.2. ISSR and SCoT marker analysis
Agarose gels containing PCR products of different ISSR and SCoT primers were carefully visualized for analyzing variations among 52 T. dioica accessions (Figs. S2, S3). A total of 127 bands were produced by 8 ISSR primers out of which 122 bands were found to be polymorphic with 95.96% polymorphism (Table 2). Mean value of PIC, EMR, MI and RP was 0.47, 14.68, 6.97 and 18.05, respectively (Table 2). Among different ISSR primers tested, VIS20 revealed maximum PIC value of 0.496, VIS11 produced highest EMR (18) and MI (8.91) values and UBC834 showed maximum RP value (21.23) (Table 2).
Nine SCoT primers produced a total of 123 bands, out of which 113 bands were found to be polymorphic (92.20% polymorphism) (Table 3). Mean of PIC, EMR, MI and RP was 0.45, 11.66, 5.33 and 17.11, respectively (Table 3). Among different SCoT primers used, SCoT34 revealed maximum PIC value of 0.496, SCoT40 produced maximum values of EMR (17) and MI (8.41) and highest RP value of 22 is shown by SCoT 28 (Table 3).
3.1.3. Population genetic diversity analysis
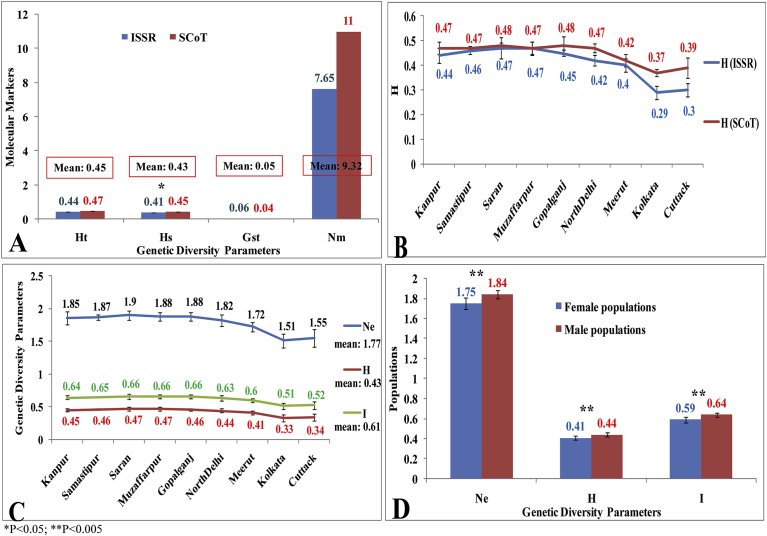
Mean total genetic diversity (Ht) and genetic diversity within populations (Hs) in T. dioica accessions collected from nine different eco-geographical regions of India was found to be very high (0.45 and 0.43 respectively). Among different T. dioica populations, very low genetic differentiation (Gst: 0.05) and high gene flow (Nm: 9.32) was observed (Fig. 1A). Ht, Hs, Gst and Nm values produced by ISSR markers were 0.44, 0.41, 0.06 and 7.65 respectively and by SCoT markers were 0.47, 0.45, 0.04, 11 respectively (Fig. 1A). Analysis of molecular variance (AMOVA) analysis revealed that 81% of the genetic variation occurred within populations and 19% of the genetic variation among populations (P: 0.001) (Table 4).
Fig. 1.
Levels of genetic diversity in different T. dioica accessions collected from different eco-geographical regions of India employing ISSR and SCoT markers using POPGENE software, (A) differences in Ht, Hs, Gst and Nm values obtained by ISSR and SCoT markers, (B) difference in 'h' value obtained by ISSR and SCoT markers among 9 different T. dioica populations, (C) differences in ne, h and I values among different T. dioica populations of India, (D) differences in ne, h and I values between female and male populations.
Table 4.
Analysis of molecular variance (AMOVA) of 52 T. dioica accessions belonging to 9 different populations.
| Source of variation | df | SS | MS | Est. Var. | % variation | P value |
|---|---|---|---|---|---|---|
| Among populations | 8 | 125.173 | 15.647 | 1.605 | 19% | 0.001 |
| Within populations | 43 | 285.250 | 6.634 | 6.634 | 81% | |
| Total | 51 | 410.423 | 8.239 | 100% |
df: degree of freedom, SS: sum of squares, MS: mean squares.
Both ISSR and SCoT markers revealed T. dioica populations of Bihar state (Samastipur, Saran, Muzaffarpur and Gopalganj) were found to be highly diverse, while populations of Kolkata and Cuttack to be least diverse (Fig. 1B). The order of genetic diversity in different populations of T. dioica was as follows; Saran (Ne: 1.90, H: 0.47, I: 0.66) ˃ Muzaffarpur (Ne: 1.88, H: 0.47, I: 0.66) ˃ Gopalganj (Ne: 1.88, H: 0.46, I: 0.66) ˃ Samastipur (Ne: 1.87, H: 0.46, I: 0.65) ˃ Kanpur (Ne: 1.85, H: 0.45, I: 0.64) ˃ North Delhi (Ne: 1.82, H: 0.44, I: 0.63) ˃ Meerut (Ne: 1.72, H: 0.41, I: 0.60) ˃ Cuttack (Ne: 1.55, H: 0.34, I: 0.52) ˃ Kolkata (Ne: 1.51, H: 0.33, I: 0.51) (Fig. 1C). Genetic diversity analysis between male and female T. dioica plants was also conducted and males were found to be more significantly genetically diverse (Ne: 1.84, H: 0.44, I: 0.64) compared to females (Ne: 1.75, H: 0.41, I: 0.59) (P < 0.05) (Fig. 1D).
3.1.4. Cluster and population genetic structure analysis
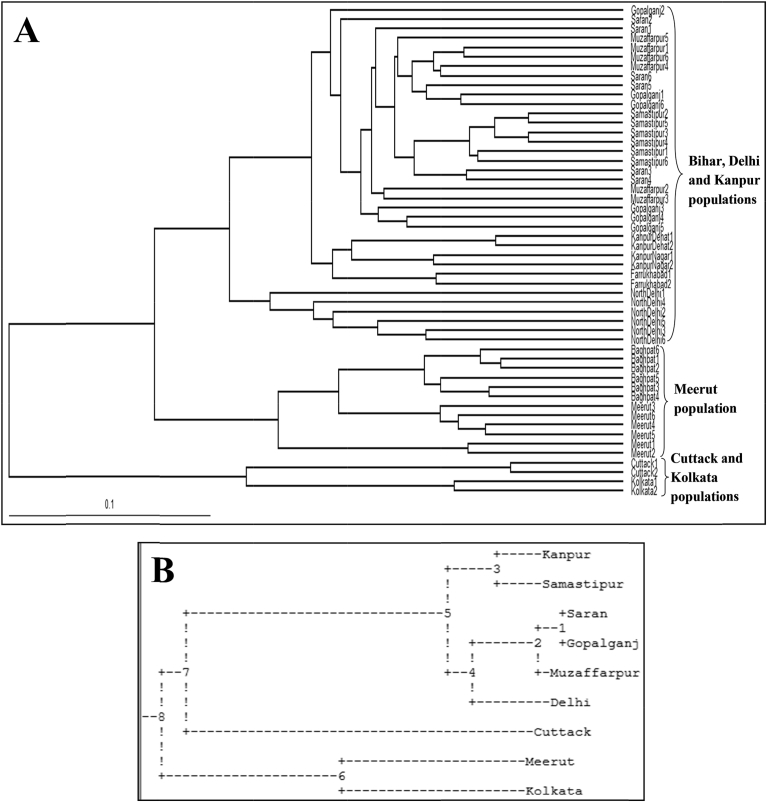
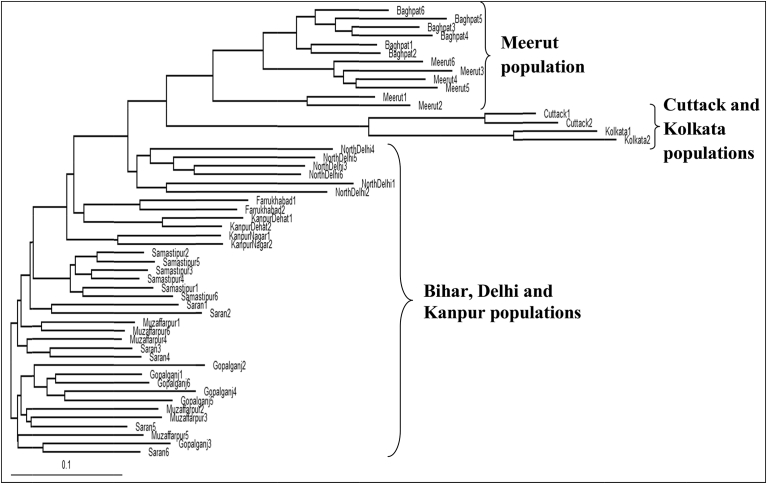
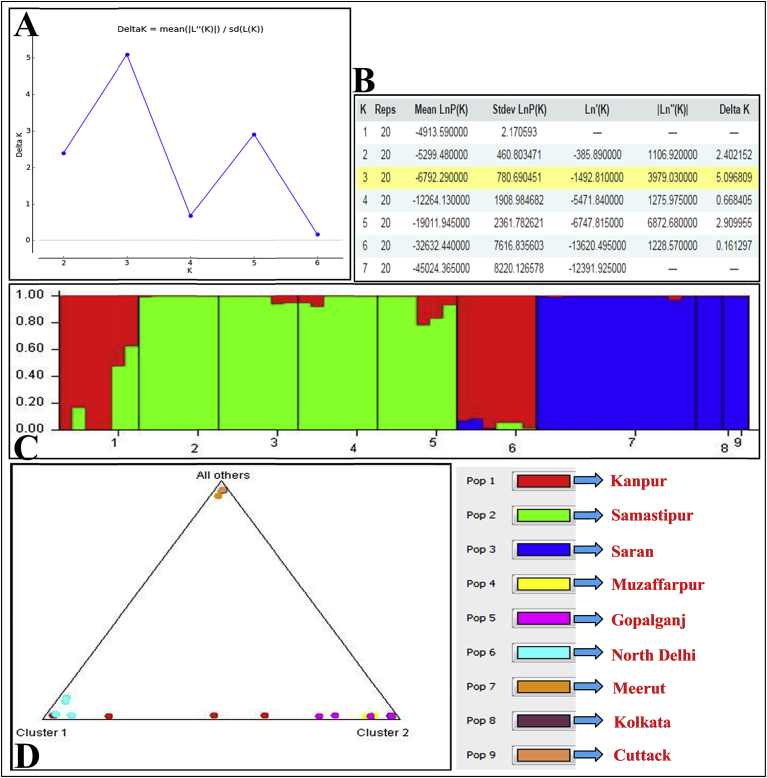
Genetic relationship among 52 T. dioica accessions was revealed by UPGMA and Neighbor-Joining trees. Both the trees divided the T. dioica accessions into three main clusters (Figs. 2 and 3). First cluster comprised of Meerut population, second cluster included Cuttack and Kolkata populations and populations of Bihar, Delhi and Kanpur occurred in third cluster (Figs. 2 and 3). T. dioica populations of Kolkata and Samastipur regions were least genetically identical (0.8766) with maximum genetic distance (0.1318) (Fig. 2B, Fig. S4). STRUCTURE software was employed to analyze genetic structure of different populations and it was run for K = 7 based on banding patterns generated by both of the markers. Based on highest ΔK value generated by STRUCTURE HARVESTER software, the number of optimum clusters was found to be three (Figs. 4A, B, Fig. S5). Cluster one comprised mainly the accessions of Delhi, Kanpur, cluster two consisted accessions of Samastipur, Saran, Muzaffarpur and Gopalganj and cluster three contained accessions of Meerut, Kolkata and Cuttack (Figs. 4C–D).
Fig. 2.
(A) UPGMA tree of 52 T. dioica accessions based on ISSR and SCoT markers using Tree View software based on Jaccard's coefficient of similarity, (B) UPGMA tree based on Nei's genetic distance, generated by POPGENE program.
Fig. 3.
Neighbor Joining tree of 52 T. dioica accessions based on ISSR and SCoT markers using Tree View software based on Jaccard's coefficient of similarity.
Fig. 4.
Population structure of different T. dioica accessions in India using STRUCTURE software version 2.3.4 and Structure Harvester v6.0 program, (A) ΔK values for different numbers of populations assumed (K) in the STRUCURE analysis, (B) Evanno table output, (C) bar plot representation for K = 3, (D) triangle plot representation for K = 3.
3.1.5. Correlation between the genetic diversity parameters and environmental factors of different T. dioica populations
Effective number of alleles (Ne), Nei's gene diversity (H) and Shannon's information index (I) were found to be significantly and strongly positively correlated with the latitude (r: 0.67, 0.69 and 0.70 respectively), and strongly negatively correlated with the mean annual rainfall (r: -0.68, -0.69 and -0.69 respectively) of different T. dioica cultivated regions. Moderate negative correlation was observed between different genetic diversity parameters (Ne, H and I) with mean minimum temperature (r: -0.55, -0.55 and -0.57 respectively) of different T. dioica populations, however, the correlation was not significant (P > 0.05) (Table 5).
Table 5.
Correlation between genetic diversity parameters (Ne, H and I) and environmental factors of T. dioica populations.
| Genetic diversity parameters | Latitude | Longitude | Elevation | Mean maximum temperature | Mean minimum temperature | Annual rainfall |
|---|---|---|---|---|---|---|
| Ne | 0.671187* | −0.28468 | 0.172675 | −0.38497 | −0.54961 | −0.68193* |
| H | 0.687475* | −0.28495 | 0.182636 | −0.36757 | −0.55323 | −0.69084* |
| I | 0.700232* | −0.29548 | 0.194491 | −0.35758 | −0.57309 | −0.69346* |
*P < 0.05; **P < 0.005.
3.2. For identification of sex linked molecular markers
3.2.1. BSA- SRAP marker analysis
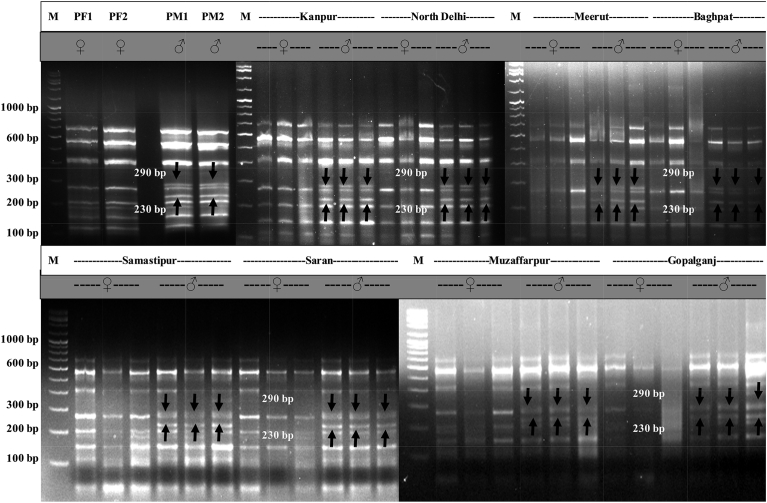
Out of total 50 SRAP primer combinations tried, one primer combination 'Em-6/Me-4' amplified two male specific bands of ∼230 and ∼290 bp in the pooled DNA samples (Fig. 5).
Fig. 5.
Male sex specific markers of size ∼290 and ∼230 bp amplified by SRAP primer combination 'Em-6/Me-4' in different T. dioica plants growing in Kanpur, North Delhi, Meerut, Baghpat, Samastipur, Saran, Muzaffarpur and Gopalganj regions. The arrows indicate male specific bands of ∼290 and ∼230 bp sizes present in only male bulk and male individuals. M: 3 kb ladder, PF: Pooled female, PM: Pooled male.
3.2.2. Validation of the putative sex linked SRAP markers on larger populations
Primer combination 'Em-6/Me-4' yielded consistent bands of ∼230 and ∼290 bp (sex linked markers) in all the male plants of Kanpur, Meerut, Baghpat and Bihar (Samastipur, Saran, Muzaffarpur and Gopalganj) regions, while were entirely absent from the female plants (Fig. 5).
4. Discussion
4.1. ISSR and SCoT marker analysis
In Trichosanthes dioica, genetic diversity and sex identification studies are very valuable as farmers prefer vegetative method for its propagation in the desire of obtaining more fruits and inability of sex identification in seeds raised plants prior flowering. However, vegetative propagation may result in erosion of genetic diversity, which is the key of sustainable high yields. Thus, the present study was conducted to analyze the genetic diversity among fifty two T. dioica accessions belonging to nine different eco-geographical regions of India employing ISSR and SCoT markers. ISSR markers proved to be more efficient in genetic diversity assessment and produced more number of polymorphic bands (122), percentage polymorphism (95.96%), PIC (0.47), EMR (14.68), MI (6.97) and RP (18.05) values compared to SCoT markers (113, 92.20%, 0.45, 11.66, 5.33 and 17.11, respectively). The efficiency of both ISSR and SCoT markers has been tested and compared in different studies. Similar to our results, higher mean of PIC (0.41), RP (13.81) and MI (4.25) were produced by ISSR primers than SCoT primers (PIC: 0.39, P: 10.96 and MI:3.74), however, SCoT primers produced 100% polymorphism and ISSR primers detected 93.61% polymorphism in Triticum turgidum var. durum in a study conducted by Etminan et al [32]. In Cucurbita pepo, Xanthopoulou et al [33] observed higher PIC value of 0.237 produced by ISSR markers compared to SCoT markers (PIC: 0.207). However in the same study, SCoT markers produced higher percentage polymorphism (62.82%), RP (4.595) and MI (1.503) compared to ISSR markers (60.50%, 3.350 and 0.965 respectively). Gorji et al [34] also observed higher PIC value of 0.210 produced by ISSR primers in comparison to SCoT primers (PIC: 0.181) in tetraploid potato genotypes, however in varieties of tetraploid potato, SCoT primers produced PIC value of 0.40 and ISSR primers produced PIC value of 0.34. Zhang et al [35] observed that SCoT markers produced percentage polymorphism of 0.90, EMR (20.10) and MI (8.64), while ISSR markers detected 0.89% polymorphism, EMR (12.25) and MI (4.66) in Panicum virgatum L. In Lycopersicum esculentum Mill., while SCoT primers produced percentage polymorphism of 36.14%, PIC (0.142) and RP (1.88), ISSR markers detected only 23.25%, PIC (0.088) and RP (1.55) [36].
4.2. Population genetic diversity analysis
Total genetic diversity (Ht) and genetic diversity within populations (Hs) was found to be very high (0.45 and 0.43 respectively) in T. dioica accessions collected from nine different eco-geographical regions of India. AMOVA analysis also revealed higher genetic variation within populations (81%) than among populations (19%). Different factors such as mating system, gene flow, population size, selection, etc. influence genetic diversity levels within populations. High genetic diversity in T. dioica accessions could be explained on the basis of its out-crossing mode of reproduction with cross pollination, perennial nature, nativity, etc. Mating system of any plant species have significant impact on genetic diversity levels [37]. Out-crossing plant species keep together new genes combinations rapidly and commonly have high genetic diversity. Long lived perennial plant species are considered to be more genetically diverse than short lived perennials [38]. Though T. dioica can reproduce sexually through seeds, this plant is mainly cultivated through vine cuttings and root suckers by farmers due to poor seed germination and inability to determine sex of the seeds raised plants prior flowering. However, asexual reproduction results in keeping together old genes combination, may result in low genetic diversity. In T. dioica, irrespective of vegetative propagation by farmers, very high genetic diversity can be explained on the basis of very high gene flow (Nm: 9.32) among different T. dioica populations, which could be due to entomophilous or anthropogenic seed or pollen dispersal to distant populations. In dioecious plants, high gene flow and low levels of genetic differentiation among populations are generally observed due to obligate outcrossing [39]. In this study also, very low genetic differentiation (Gst: 0.05) among 9 T. dioica populations was observed. Similar to our results, high levels of genetic diversity in different dioecious plants has also been observed in other studies. In Antennaria dioica, the high levels of genetic diversity (He: 0.18–0.22), low genetic differentiation (ΦST: 0.11) and higher molecular variance of 89% within populations than among populations (11%) were observed employing AFLP markers [40]. Zhai et al [41] observed high genetic diversity (He: 0.7566), moderate differentiation (FST: 0.0858) and high genetic variation within subpopulations (94%) in dioecious shrub Salix viminalis employing 20 SSR loci. Dev et al [42] observed mean 'h' values of 0.6329 and 0.7677 in dioecious Ficus hispida and Ficus exasperata employing microsatellite markers. Luna et al [43] observed genetic variation in two palm species of dioecious Chamaedorea, C. tepejilote (He: 0.385–0.442) and C. elatior (He: 0.278–0.342). In dioecious plant Uapaca kirkiana, AFLP markers revealed moderate differentiation (GST: 0.079) and higher genetic variation (92%) among different individuals within eight populations than among populations (6.8%) [44]. Terauchi [45] observed a high level of genetic diversity (HT: 0.282) in dioecious climber, Dioscorea tokoro Makino employing allozymes.
Origin of any plant species is also an important factor in retaining high genetic diversity levels, as introduced populations have higher risks of founder effects resulting in low genetic diversity. T. dioica is native to Indian sub-continent and this could be the probable reason for having high genetic diversity in different populations of this plant in India. Similar to our results, Shirk et al [46] observed high genetic diversity Geranium carolinianum (annual herb) in its native place of North America compared to its introduced populations in China due to founder effects. Higher genetic diversity in thirty eight native populations of Capsicum annuum L. var. annuum belonging to seven states of Mexico, was revealed by twenty four microsatellite loci [47].
Different populations have the different genetic diversity levels and identification of populations with low genetic diversity is very important for making management strategies. Bengal-Assam regions are the primary centre of origin of T. dioica [48]. and from there, this plant dispersed to other regions of India such as Bihar, West-Bengal, Eastern parts of Uttar Pradesh, Assam, Odisha, Madhya Pradesh, etc. Genetic structure of introduced populations could differ from native population due to genetic drift, local adaptation and founder effect processes. During current study, T. dioica populations of Bihar state was found to be highly polymorphic and population of Kolkata (West Bengal) was found to be least variable. Differences in genetic diversity levels among different populations could be attributed to different cultivation patterns (different cultivars and mode of cultivation), change in environmental conditions, etc. High genetic diversity levels in introduced populations than native populations due to high propagule pressure has also been observed by Genton et al [49] and Kolbe et al [50]. Similar to molecular diversity, morphological variations among different T. dioica accessions belonging to nine different populations were found to be very high. Similar to our results, Bharathi [51] found high phenotypic variations among 22 different T. dioica landraces. T. dioica populations of Bihar state were of more desired traits (early flowering, fruits length and weight) in comparison to other populations. The reason could be attributed to its cultivation style of training on trellis system for aerial support, which results in luxuriant plant growth and earlier flowering [5].
In dioecious plants, genetic diversity assessment in female and male plants is very important for understanding of evolutionary processes of a species and making successful breeding programs. During present study, male plants were found to be more genetically polymorphic in comparison to females. Differences in genetic diversity levels between sexes could be due to various factors such as sex ratio, individuals distribution patterns, stochastic events, etc. Reproductive systems and history of species have direct influence on the levels and distribution of genetic variation, genetic divergence, and genetic structure [52, 53, 54, 55, 56, 57]. Similar to our results, Heikrujam et al [58] also observed high genetic diversity in male Simmondsia chinensis genotypes over female genotypes employing SCoT and CBDB markers. Higher genetically variable male populations (H: 0.252 and I: 0.387) than female populations (H: 0.249 and I: 0.381) were also observed in dioecious Pistacia atlantica by Nosrati et al [59]. Ranade et al [60] also observed more genetic diversity among male plants than female plants in dioecious Piper betle. However, Kumar and Agrawal [61] observed equal amount of genetic diversity in male and female plants of Simarouba glauca.
4.3. Cluster and population genetic structure analysis
UPGMA and NJ trees were constructed employing combined data of ISSR and SCoT markers for analyzing genetic relationships among different T. dioica accessions in India. Both trees grouped T. dioica accessions into three clusters. First cluster comprised of Meerut population, second cluster included Cuttack and Kolkata populations and populations of Bihar, Delhi and Kanpur occurred in third cluster. This classification of T. dioica accessions could be explained on the basis of differences in eco-geographical characteristics of different populations. T. dioica populations of Kolkata and Cuttack regions were found to be least similar to other populations. The extremely different geographical coordinates and climatic conditions (low altitude, high rainfall, least difference in mean minimum and mean maximum temperature) could be the probable reason. Similarly, Meerut region is situated at higher altitude of around 225 meters compared to other regions. Applicability of ISSR and SCoT markers in grouping genetically similar individuals has also been observed in Salvia [16], Vigna unguiculata [62], Cicer arietinum [63], etc. Similar to UPGMA and NJ trees obtained in the current study, grouping of T. dioica accessions into three cluster was also revealed by STRUCTURE analysis, one comprised mainly the accessions of Delhi, Kanpur, cluster two consisted accessions of Samastipur, Saran, Muzaffarpur and Gopalganj and cluster three contained accessions of Meerut, Kolkata and Cuttack. Admixture of different T. dioica accessions was also observed and could be the result of seed dispersal by humans. STRUCTURE software distributes different accessions to different populations depending upon allele frequencies of the accessions. Differences in ecological pressures and ecological factors among different populations could be attributed to differences in genetic structure and distribution of genetic variations [64, 65]. Similar to our results, clustering based on geographical regions has also been seen in different crop plants such as Afzelia xylocarpa [66], Arabidopsis thaliana [67], Triticum aestivum [68], Pulsatilla patens [69], etc.
4.4. Correlation between the genetic diversity parameters and environmental factors of different T. dioica populations
Change in environmental conditions, such as difference in temperature, rainfall, etc. is also expected to alter genetic diversity levels among different populations [10, 70]. During present study, Genetic diversity was found to strongly positively correlated with the latitude and strongly negatively correlated with mean annual rainfall of different T. dioica cultivated regions and a moderate negative correlation was observed between 'H' and mean maximum and mean minimum temperature. A weak negative correlation was observed between 'H' and longitude and weak positive correlation was found between 'H' and elevation. Similar to our results, several studies have revealed correlation of environmental factors with genetic diversity. Hu et al [71] observed a positive correlation of genetic diversity of Rheum tanguticum with annual mean precipitation and altitude and negative correlation with annual mean temperature and latitude. Schoettle et al [72] observed positive correlation between genetic diversity of Pinus aristata and longitude and elevation of different populations. Shen et al [73] observed no clear correlation of genetic variation with altitude in Populus szechuanica. Genetic diversity of Caragana microphylla was found to be positively correlated with mean annual rainfall and negatively associated with annual mean temperature [9].
4.5. For identification of sex linked molecular markers
In T. dioica, mostly female plants are preferred as the plant is cultivated mainly for its fruits. However, sex identification in seeds raised plants is not possible prior flowering which takes around 7–8 months. A molecular method to identify sex of the plant at early seedling stages will encourage farmers to propagate T. dioica through seeds. During current investigation, one SRAP primer combination, 'Em-6/Me-4' produced two male specific bands of ∼230 and ∼290 bp in pooled and individual samples of Bihar, Kanpur, North Delhi, Meerut and Baghpat regions. These sex linked molecular markers were completely absent from female samples. These sex linked SRAP markers could be very useful in distinguishing male plants from the females at early seedling stages in this economically important crop and thus will save time, money and resources of the farmers. Prior to our study, SRAP markers have been used for sex identification only in few plants. Female sex specific SRAP markers have been identified in Buchloe dactyloides [20], Idesia polycarpa [21], Ficus pumila [74] and Litsea cubeba [75].
5. Conclusions
Our results revealed a high level of genetic diversity and gene flow in T. dioica accessions belonging to nine different populations with different eco-geographical characteristics. Both ISSR and SCoT markers proved to be efficient in assessing genetic variations in different populations. T. dioica populations of Bihar state (Samastipur, Saran, Muzaffarpur and Gopalganj) were found to be highly polymorphic and population of Kolkata was least diverse and needs to be conserved. Genetic diversity was found to be correlated with different environmental factors. Male plants displayed higher genetic diversity levels than female plants. Cluster analysis divided T. dioica accessions into three main clusters based on differences in eco-geographical characteristics of different populations. For sex identification prior flowering, two male specific markers of ∼230 and ∼290 bp were identified, which were produced by SRAP primer combination, 'Em-6/Me-4'. These markers were positively validated on larger populations of Bihar, Kanpur, North Delhi, Meerut and Baghpat regions.
Declarations
Author contribution statement
Jatin Kumar: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Veena Agrawal: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Science and Engineering Research Board, Government of India, New Delhi which sanctioned the Major Research Projects (SERB/SR/SO/PS/05/2012 and EMR/2016/001673) to Prof Veena Agrawal; the UGC RGNF fellowship which was provided to Jatin Kumar; and the University of Delhi, India for providing Research and Development Grant.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Choudhury B. National Book Trust.; New Delhi: 1979. Vegetables; pp. 160–162. [Google Scholar]
- 2.Haines H.H. reprinted edition. Vol II. Botanical Survey and India; Calcutta: 1961. p. 406. (The Botany of Bihar and Orissa). [Google Scholar]
- 3.Hooker J.D. Reprinted edition. Periodical experts; Delhi: 1973. Flora of British India. pp. II 609. [Google Scholar]
- 4.Kanjilal V.N. Reprinted edition. Vol. II. 1997. p. 329. (Flora of Assam). [Google Scholar]
- 5.Singh K. Pointed gourd (Trichosanthes dioica Roxb.) Indian Hortic. 1989:34–37. [Google Scholar]
- 6.Som M.G., Maity T.K., Hazra P. Pointed gourd Trichosanthes dioica Roxb. In: Kalloo G., Berg B.O., editors. Genetic Improvement of Vegetable Crops. Pergamon Press; Oxford, UK: 1993. pp. 251–258. [Google Scholar]
- 7.Kumar S., Singh B.D., Pandey S., Ram D. Inheritance of leaf and stem morphological traits in pointed gourd (Trichosanthes dioica Roxb.) J. Crop. Improv. 2008;22:225–233. [Google Scholar]
- 8.Quinones-Perez C.Z., Saenz Romero C., Wehenkel C. Genetic diversity and conservation of Picea chihuahuana Martinez: a review. Afr. J. Biotechnol. 2014;13:2786–2795. [Google Scholar]
- 9.Huang W., Zhao X., Zhao X., Li Y., Lian J. Effects of environmental factors on genetic diversity of Caragana microphylla in Horqin sandy land, northeast China. Ecol. Evol. 2016;6:8256–8266. doi: 10.1002/ece3.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovejoy T.E., Hannah L.J. Yale University Press; New Haven, CT: 2005. Climate Change and Biodiversity. [Google Scholar]
- 11.Hilfiker K., Gugerli F., Schutz J.P., Rotach P.A., Holderegger R. Low RAPD variation and female-biased sex ratio indicate genetic drift in small populations of the dioecious conifer Taxus baccata in Switzerland. Conserv. Genet. 2004;5:357–365. [Google Scholar]
- 12.Vandepitte K., Honnay O., De Meyer T., Jacquemyn H., Roldan-Ruiz I. Patterns of sex ratio variation and genetic diversity in the dioecious forest perennial Mercurialis perennis. Plant Ecol. 2010;206:105–114. [Google Scholar]
- 13.De Jong T.J., Klinkhamer P.G.L. Cambridge University Press; Cambridge, U.K: 2005. Evolutionary Ecology of Plant Reproductive Strategies. [Google Scholar]
- 14.Engen S., Lande R., Saether B.E. Demographic stochasticity and allee effects in populations with two sexes. Ecology. 2003;84:2378–2386. [Google Scholar]
- 15.Khanam S., Sham A., Bennetgen J.L., Mohammed A.M.A. Analysis of molecular marker-based characterization and genetic variation in date palm (Phoenix dactylifera L.), Austral. J. Crop Sci. 2012;6:1236–1244. [Google Scholar]
- 16.Etminan A., Pour-Aboughadareh A., Noori A., Ahmadi-Rad A., Shooshtari L., Mahdavian Z., Yousefiazar-Khanian M. Genetic relationships and diversity among wild Salvia accessions revealed by ISSR and SCoT markers. Biotechnol. Biotechnol. Equip. 2018;32:610–617. [Google Scholar]
- 17.Agarwal A., Gupta V., Haq S.U., Jatav P.K., Kothari S.L., Kachhwaha S. Assessment of genetic diversity in 29 rose germplasms using SCoT marker. JKSUS. 2018 [Google Scholar]
- 18.Abdel-Lateif K.S., Hewedy O.A. Genetic diversity among Egyptian wheat cultivars using SCoT and ISSR markers. SABRAO J. Breed. Genet. 2018;50:36–45. [Google Scholar]
- 19.Rajalakshmi R., Rajalakshmi S., Parida A. Genetic diversity, population structure and correlation study in Moringa oleifera Lam. using ISSR and SRAP markers. Proc. Natl. Acad. Sci. India B Biol. Sci. 2018:1–11. [Google Scholar]
- 20.Zhou Y.J., Wang X.G., Zhang X.Q. Development and application of a SRAP marker for the identification of sex in Buchloe dactyloides. Euphytica. 2011;181:261–266. [Google Scholar]
- 21.Wang S.H., Li Y., Li Z.Q., Chang L., Li L. Identification of an SCAR marker related to female phenotype in Idesia polycarpa Maxim. Genet. Mol. Res. 2015;14:2015–2022. doi: 10.4238/2015.March.20.11. [DOI] [PubMed] [Google Scholar]
- 22.Saghai-Maroof M.A., Soliman K.M., Jorgensen R.A., Allard R.W. Ribosomal DNA spacer-length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. 1984;81:8014–8019. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith J.S.C., Chin E.C., Shu H., Smith O.S., Wall S.J., Senior M.L., Mitchell S.E., Kresovich S., Ziegle J. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.) comparisons with data from RFLPs and pedigree. Theor. Appl. Genet. 1997;95:163–173. [Google Scholar]
- 24.Yeh Francis C., Yang R.C., Boyle Timothy B.J., Ye Z.H., Mao Judy X. University of Alberta; Canada: 1999. POPGENE Version 1.32, the User-Friendly Shareware for Population Genetic Analysis, Molecular Biology and Biotechnology Centre. [Google Scholar]
- 25.Pavlicek A., Hrda S., Flegr J. Free Tree: freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol. (Praha) 1999;45:97–99. [PubMed] [Google Scholar]
- 26.Page R.D. Tree View: an application to display phylogenetic trees on personal computers. Comput. Mol. Biol. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 29.Earl Dent A., vonHoldt Bridgett M. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. [Google Scholar]
- 30.Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelmore R.W., Paran I., Kesseli R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etminan A., Pour-Aboughadareh A., Mohammadi R., Ahmadi-Rad A., Noori A., Mahdavian Z., Moradi Z. Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol. Biotechnol. Equip. 2016;30:1075–1081. [Google Scholar]
- 33.Xanthopoulou A., Ganopoulos I., Kalivas A., Nianiou-Obeidat I., Ralli P., Moysiadis T., Tsaftaris A., Madesis P. Comparative analysis of genetic diversity in Greek Genebank collection of summer squash ('Cucurbita pepo') landraces using start codon targeted (SCoT) polymorphism and ISSR markers. Aust. J. Crop. Sci. 2015;9:14–21. [Google Scholar]
- 34.Gorji A.M., Poczai P., Polgar Z., Taller J. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am. J. Potato Res. 2011;88:226–237. [Google Scholar]
- 35.Zhang Y., Yan H., Jiang X., Wang X., Huang L., Xu B., Zhang X., Zhang L. Genetic variation, population structure and linkage disequilibrium in Switchgrass with ISSR, SCoT and EST-SSR markers. Hereditas. 2016;153:4. doi: 10.1186/s41065-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahlaei A., Torabi S., Khosroshahli M. Efficacy of SCoT and ISSR markers in assessment of tomato (Lycopersicum esculentum Mill.) genetic diversity. Int. J. Biosci. 2014;5:14–22. [Google Scholar]
- 37.Charlesworth D., Wright S.I. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- 38.Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- 39.Hamrick J.L., Godt M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. Roy. Soc. London Sect. B. 1996;351:1292–1298. [Google Scholar]
- 40.Rosche C., Schrieber K., Lachmuth S., Durka W., Hirsch H., Wagner V., Schleuning M., Hensen I. Sex ratio rather than population size affects genetic diversity in Antennaria dioica. Plant Biol. 2018;20:789–796. doi: 10.1111/plb.12716. [DOI] [PubMed] [Google Scholar]
- 41.Zhai F., Mao J., Liu J., Peng X., Han L., Sun Z. Male and female subpopulations of Salix viminalis present high genetic diversity and high long-term migration rates between them. Front. Plant Sci. 2016;7:330. doi: 10.3389/fpls.2016.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dev S.A., Kjellberg F., Hossaert-McKey M., Borges R.M. Fine-scale population genetic structure of two dioecious Indian keystone species, Ficus hispida and Ficus exasperata (Moraceae) Biotropica. 2011;43:309–316. [Google Scholar]
- 43.Luna R., Epperson B.K., Oyama K. High levels of genetic variability and inbreeding in two neotropical dioecious palms with contrasting life histories. Heredity. 2007;99:466–476. doi: 10.1038/sj.hdy.6801027. [DOI] [PubMed] [Google Scholar]
- 44.Mwase W.F., Bjornstad A., Stedje B., Bokosi J.M., Kwapata M.B. Genetic diversity of Uapaca kirkiana Muel. Arg. populations as revealed by amplified fragment length polymorphisms (AFLPs) Afr. J. Biotechnol. 2006;5:1205–1213. [Google Scholar]
- 45.Terauchi R. Genetic diversity and population structure of Dioscorea tokoro Makino, a dioecious climber. Plant Species Biol. 1990;5:243–253. [Google Scholar]
- 46.Shirk R.Y., Hamrick J.L., Zhang C., Qiang S. Patterns of genetic diversity reveal multiple introductions and recurrent founder effects during range expansion in invasive populations of Geranium carolinianum (Geraniaceae) Heredity. 2014;112:497–507. doi: 10.1038/hdy.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toledo-Aguilar R., Lopez-Sanchez H., Santacruz-Varela A., Valadez-Moctezuma E., Lopez P.A., Aguilar-Rincon V.H., Gonzalez-Hernandez V.A., Vaquera-Huerta H. Characterization of genetic diversity of native 'Ancho' chili populations of Mexico using microsatellite markers. Chilean J. Agric. Res. 2016;76:18–26. [Google Scholar]
- 48.Singh A.K., Singh R.D., Singh K. Genetic variability, heritability and genetic advance for some traits in pointed gourd (Trichosanthes dioica Roxb.) Haryana J. Hort. Sci. 1992;21:236–240. [Google Scholar]
- 49.Genton B.J., Shykoff J.A., Giraud T. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 2005;14:4275–4285. doi: 10.1111/j.1365-294X.2005.02750.x. [DOI] [PubMed] [Google Scholar]
- 50.Kolbe J.J., Glor R.E., Rodriguez Schettino L., Lara A.C., Larson A., Losos J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- 51.Bharathi L.K. Phenotypic diversity analysis in pointed gourd (Trichosanthes dioica Roxb.) Cucurbit Genet. Coop. 2010;33/34:62–64. [Google Scholar]
- 52.Loveless M.D., Hamrick J.L. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst. 1984;15:65–95. [Google Scholar]
- 53.Hamrick J.L., Godt J.W. Allozyme diversity in plant species. In: Brown A.H.D., Clegg M.T., Kahler A.L., Weir B.S., editors. Plant Population Genetics, Breeding, and Genetic Resources. Sin-auer Associates; Sunderland, Massachusetts: 1989. pp. 43–63. [Google Scholar]
- 54.Karron J.D. Patterns of genetic variation and breeding systems in rare plant species. In: Falk D.A., Holsinger K.E., editors. Genetics and Conservation of Rare Plants. Oxford University Press; New York: 1991. [Google Scholar]
- 55.Charlesworth D., Charlesworth B. Quantitative genetics in plants: the effect of the breeding system on genetic variability. Evol. 1995;49:911–920. doi: 10.1111/j.1558-5646.1995.tb02326.x. [DOI] [PubMed] [Google Scholar]
- 56.Hamrick J.L., Godt M.J.W. Effects of life history traits on genetic diversity in plant species. In: Silvertown J., Franco M., Harper J.L., editors. Plant Life Histories: Ecology, Phylogeny, and Evolution. Cambridge University Press; Cambridge: 1997. [Google Scholar]
- 57.Segarra-Moragues J.G., Catalan P. Low allozyme variabilityin the critically endangered Borderea chouardii and in its congener Borderea pyrenaica (Dioscoreaceae), two palaeoendemicrelicts from the central Pyrenees. Int. J. Plant Sci. 2002;163:159–166. [Google Scholar]
- 58.Heikrujam M., Kumar J., Agrawal V. Genetic diversity analysis among male and female Jojoba genotypes employing gene targeted molecular markers, start codon targeted (SCoT) polymorphism and CAAT box-derived polymorphism (CBDP) markers. Meta gene. 2015;5:90–97. doi: 10.1016/j.mgene.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nosrati H., Husainpourfeizi M.A., Khorasani M., Razban-Haghighi A., Nikniazi M. Sex ratio and genetic diversity in the dioecious Pistacia atlantica (Anacardiaceae) J. Agrobiol. 2012;29:41–46. [Google Scholar]
- 60.Ranade S.A., Soni A., Kumar N. SPAR Profiles for the assessment of genetic diversity between male and female landraces of the dioecious betelvine plant (Piper betle L.) In: Grillo O., editor. Biodiversity-Book 1. Intech Open Access Publishers; Croatia: 2011. [Google Scholar]
- 61.Kumar J., Agrawal V. Analysis of genetic diversity and population genetic structure in Simarouba glauca DC. (an important bio-energy crop) employing ISSR and SRAP markers. Ind. Crops Prod. 2017;100:198–207. [Google Scholar]
- 62.Igwe D.O., Afiukwa C.A., Ubi B.E., Ogbu K.I., Ojuederie O.B., Ude G.N. Assessment of genetic diversity in Vigna unguiculata L.(Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Genet. 2017;18:98. doi: 10.1186/s12863-017-0567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pakseresht F., Talebi R., Karami E. Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants. 2013:563–574. doi: 10.1007/s12298-013-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baatout H., Marrakchi M., Pernes J. Electrophoretic studies of genetic variation in natural populations of allogamous Hedysarum capitatum and autogamous H. euspinosissimum. Plant Sci. 1990;69:49–64. [Google Scholar]
- 65.Al-Hiyaly S.E.K., McNeilly T., Bradshaw A.D. The effect of zinc contamination from electricity pylons. Genetic constraints on selection for zinc tolerance. Heredity. 1993;70:22–32. [Google Scholar]
- 66.Pakkad G., Kanetani S., Elliott S. Genetic diversity and differentiation of an endangered tree species, Afzelia xylocarpa (Kurz) craib in Thailand revealed by nuclear microsatellite markers. Afr. J. Biotechnol. 2014;13:366–377. [Google Scholar]
- 67.Tyagi A., Singh S., Mishra P., Singh A., Tripathi A.M., Jena S.N., Roy S. Genetic diversity and population structure of Arabidopsis thaliana along an altitudinal gradient. AoB Plants. 2015;8:plv145. doi: 10.1093/aobpla/plv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan M.K., Pandey A., Thomas G., Akkaya M.S., Kayis S.A., Ozsensoy Y., Hamurcu M., Gezgin S., Topal A., Hakki E.E. Genetic diversity and population structure of wheat in India and Turkey. AoB Plants. 2015;7:plv083. doi: 10.1093/aobpla/plv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szczecinska M., Sramko G., Wolosz K., Sawicki J. Genetic diversity and population structure of the rare and endangered plant species Pulsatilla patens (L.) mill in east central Europe. PLoS One. 2016;11:e0151730. doi: 10.1371/journal.pone.0151730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keller S.R., Soolanayakanahally R., Guy R.D., Silim S.N., Olson M., Tiffin P. Climate-driven local adaptation of ecophysiology and phenology in balsam poplar, Populus balsamifera L. (Salicaceae) Am. J. Bot. 2011;98:99–108. doi: 10.3732/ajb.1000317. [DOI] [PubMed] [Google Scholar]
- 71.Hu Y., Wang L., Xie X., Yang J., Li Y., Zhang H. Genetic diversity of wild populations of Rheum tanguticum endemic to China as revealed by ISSR analysis. Biochem. Syst. Ecol. 2010;38:264–274. [Google Scholar]
- 72.Schoettle A.W., Goodrich B.A., Hipkins V., Richards C., Kray J. Geographic patterns of genetic variation and population structure in Pinus aristata, Rocky Mountain bristlecone pine. Can. J. For. Res. 2011;42:23–37. [Google Scholar]
- 73.Shen D., Bo W., Xu F., Wu R. Genetic diversity and population structure of the Tibetan poplar (Populus szechuanica var. tibetica) along an altitude gradient. BMC Genet. 2014;15:S11. doi: 10.1186/1471-2156-15-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu W., Zhu X., Chen Y. Identification of a sex-associated SRAP marker in Ficus pumila L. Chin. J. Appl. Environ. Biol. 2008;5:021. [Google Scholar]
- 75.Wu Q., Chen Y., Wang Y., Lin L. Sex differential marker FD for rapid sex identification of Litsea cubeba. Genet. Mol. Res. 2015;14:12820–12827. doi: 10.4238/2015.October.21.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.