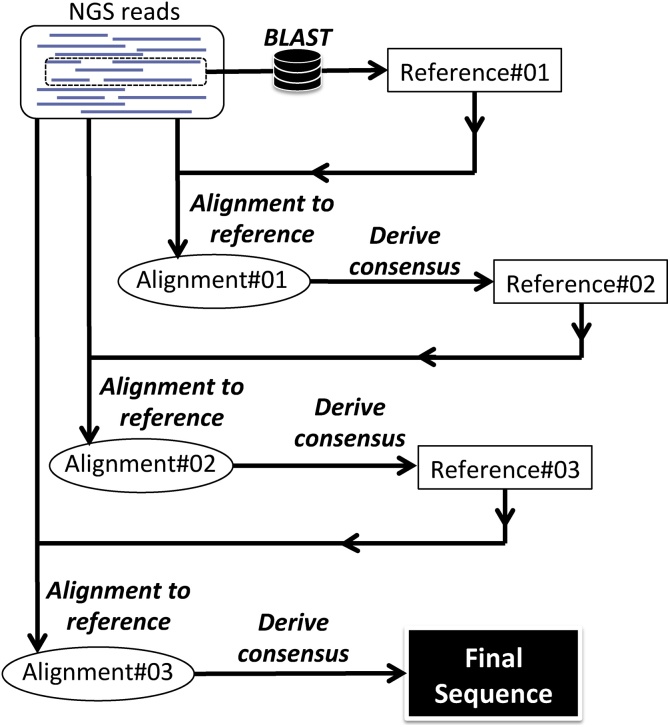
Fig. 2.
Algorithm for deriving a consensus sequence from HIV-1 single genome amplicon Using the tango software, the closest published HIV-1 sequence that can serve as a reference is obtained by applying BLAST on a random sample of NGS reads (Reference#01). The complete set of NGS reads is then aligned to Reference#01 using an implementation of BWA [45]. Afterwards, the consensus of the alignment (Reference#02) is derived by analyzing the frequency of nucleotide bases, insertions, and deletions in the sam file using Nautilus and SaGA software. Reference#02 is used to guide the alignment of NGS reads and consensus derivation, in a new iteration, until the “final consensus sequence” is obtained. See text for details.