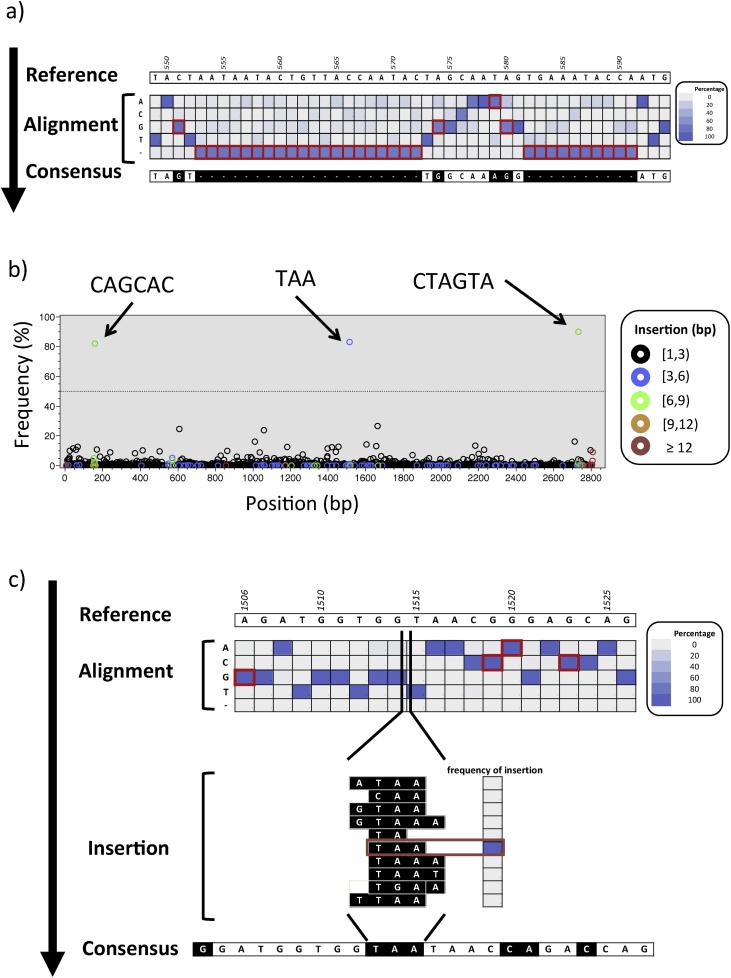
Fig. 3.
Example of derivation of single genome amplicon NGS consensus. A) At each alignment position (columns), the sequence of the reference (top) is compared with the frequency of nucleotide bases or gaps (rows) tallied based on the analysis of the sam file (to ease visualization, frequencies are here presented as a heat map). Whenever the most frequent base/gap differs from the reference (red border), the sequence of the consensus is modified accordingly (black boxes). B) By analyzing the CIGAR field in the sam alignment file it is possible to tally the sequences from the NGS reads encoded as “I”, which correspond to “insertion to the reference” (i.e., bases present in the NGS reads that do not have a corresponding position in the reference). The plot depicts, at position of the alignment (x-axis), the frequency of the insertions (y-axis). Data points are color-coded based on the length of the insertion. The arrows depict the sequences of three predominant insertions. Insertions above the operational threshold (dotted line at 50%) are followed up in downstream analysis, where C) the most common motif (in this case “TAA”) is inserted back into the new consensus sequence in the corresponding position.