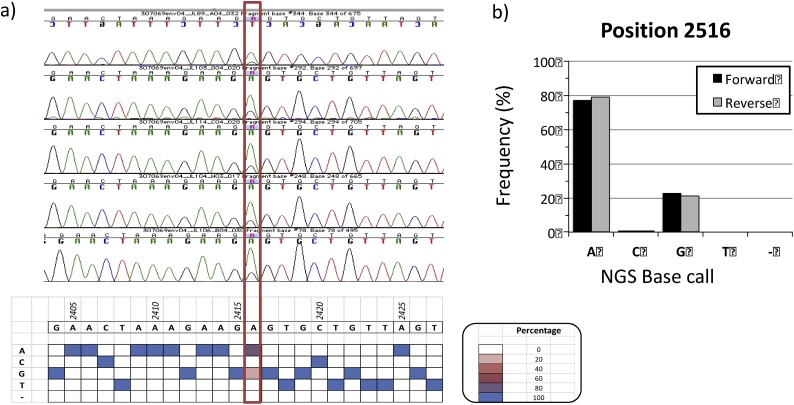
Fig. 4.
NGS-based analysis of mixed bases. A) In amplicon #04 from participant “J” 5/5 Sanger capillary sequencing chromatograms covering position 2516 (red border) show overlapping peaks for A and G, which would result in an “R” base calling based on the IUPAC nomenclature. The presence of “A” and “G” are also supported in the NGS reads (heat map on the bottom). B) NGS data shows ˜80% of reads supporting the A and 20% supporting the G, with no directional bias. Quantitation of NGS reads allows to discriminate between events that should be base called as single vs. mixed bases, in a more consistent manner.