Figure 1.
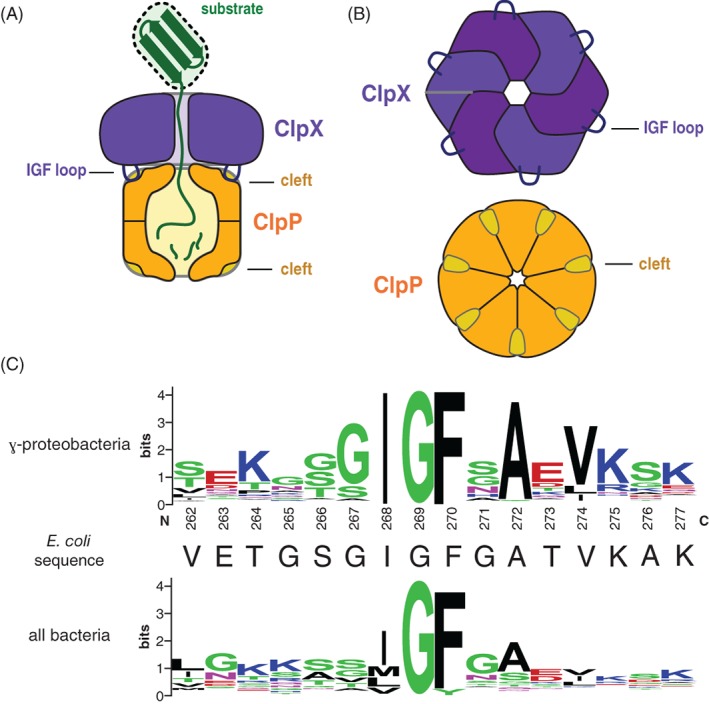
(A) Cartoon of the ClpXP protease degrading a protein substrate. The ClpX hexamer (colored light and dark purple) recognizes a protein substrate (colored green) and uses cycles of ATP hydrolysis to unfold and translocate it into the degradation chamber of the ClpP peptidase (colored dark yellow). The IGF loops of ClpX dock into hydrophobic clefts on ClpP. (B) Top views of the ClpX and ClpP rings, highlighting the six IGF loops of ClpX and seven clefts of ClpP. (C) Sequence‐logo depictions8 of sequence conservation in the IGF loops of ClpX orthologs from γ‐proteobacteria (top) and all bacteria (bottom). The sequence of the E. coli ClpX IGF loop is shown in the middle.
