Figure 2.
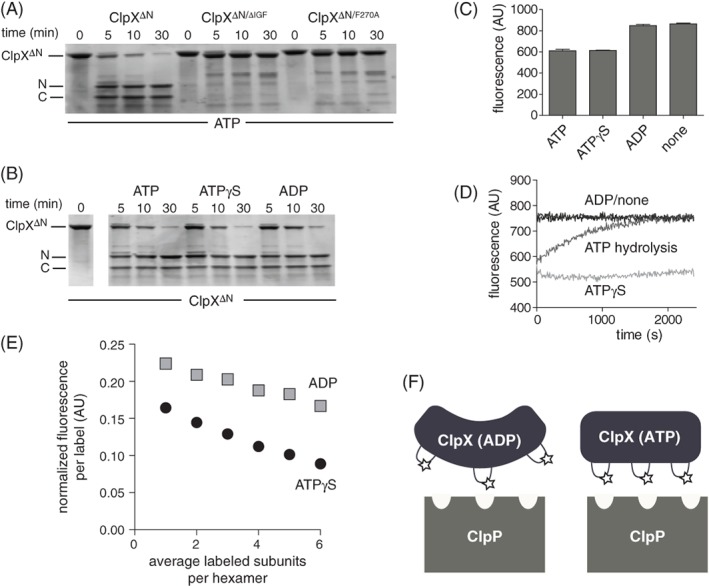
Nucleotide dependence of protease accessibility and relative distance between IGF loops. (A) As assayed by SDS‐PAGE, chymotrypsin cleaved ClpXΔN into two major fragments, labeled N and C, which were not observed following chymotrypsin incubation with ClpXΔN/ΔIGF or ClpXΔN/F270A. Experiments contained ATP (10 mM), chymotrypsin (0.01 mg/mL), and ClpXΔN variants (1 μM hexamer). (B) Chymotryptic cleavage of ClpXΔN in the presence of different nucleotides. Except for nucleotide identity, experimental conditions were the same as in Panel A. (C) Initial fluorescence of Alexa‐647 labeled T273C ClpXΔN in the presence of ATP, ATPγS, ADP, or no nucleotide. The protein concentration was 0.5 μM, and nucleotide was 1.5 mM when present. Values are averages (N = 3) ± SD. (D) Time‐dependent changes in fluorescence of Alexa‐647 labeled T273C ClpXΔN under different nucleotide conditions. Other conditions were identical to Panel C. (E) Different concentrations of unlabeled ClpXΔN and Alexa‐647 labeled T273C ClpXΔN were mixed for 1 h in the absence of nucleotide, 5 mM ADP or ATPγS was added, and fluorescence was measured. (F) Decreased fluorescence caused by increased homo quenching is consistent with the IGF loops being closer together in fluorescent T233C ClpXΔN that is bound to ATP compared to ADP. We propose that the IGF loops in ATP‐bound ClpX are properly oriented to make efficient multivalent contacts with the clefts in ClpP, whereas the IGF‐loops in ADP‐bound ClpX can only make a subset of efficient contacts.
