Figure 5.
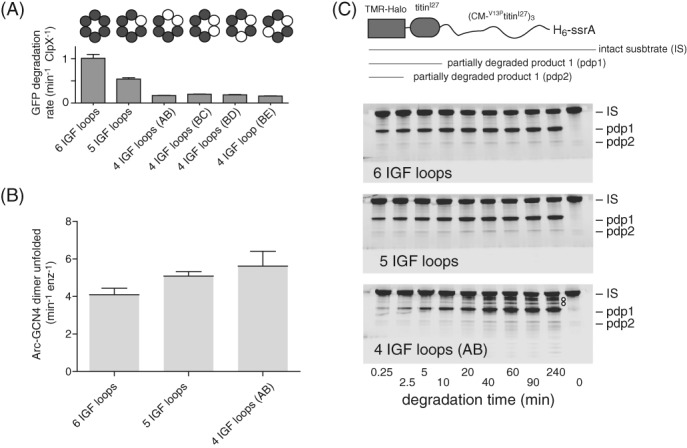
IGF‐loop deletion affects degradation rates and processivity. (A) Rates of degradation of GFP‐ssrA (20 μM) by ClpP (0.9 μM) and different variants of ClpX (0.3 μM pseudohexamer) were measured by monitoring loss of GFP fluorescence. Values are averages (N = 3) ± SD. (B) Rates of unfolding of a fluorescent Arc‐GCN4‐ssrA (5 μM) by ClpX variants (0.3 μM pseudohexamer) were measured in the presence of 10 mM ATP. (C) Top; cartoon of a substrate containing a TAMRA‐labeled Halo domain, a native titinI27 domain, three V13PtitinI27 domains unfolded by carboxymethylation (CM), and a H6‐ssrA degron. Bottom; SDS‐PAGE assays of the ClpP degradation of this substrate by ClpXΔN variants with six, five, or four IGF loops. Note that the variant with four IGF loops shows multiple additional bands between IS and pdp1, indicative of poorly processive degradation. For the enzyme with six IGF‐loops, little accumulation of the pdp1 product or loss of IS occurred after 20 min, probably because the ssrA tag is missing from the majority of substrate molecules because of exopeptidase clipping during purification. Reactions contained substrate (10 μM), ClpP (0.9 μM), single‐chain ClpXΔN variants (0.3 μM hexamer equivalents), and ATP (10 mM). Gels were imaged for fluorescence of the TAMRA dye.
