Figure 6.
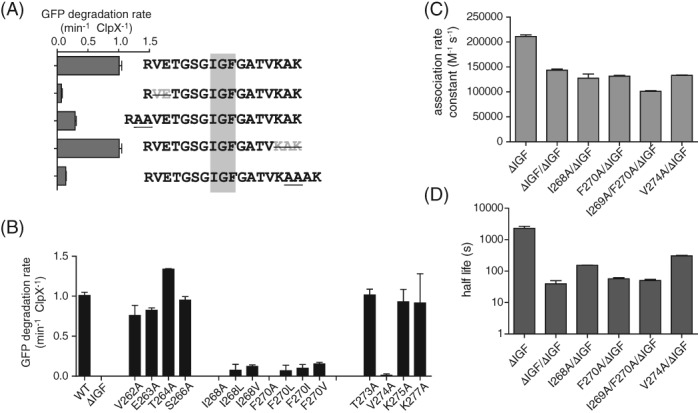
Effects of mutations in the IGF loop. (A) Rates of degradation of GFP‐ssrA (20 μM) by ClpP (0.9 μM) and ClpXΔN variants containing longer or shorter IGF loops (0.3 μM hexamer). (B) Rates of degradation of GFP‐ssrA (20 μM) by ClpP (0.9 μM) and ClpXΔN variants (0.3 μM hexamer) with single‐ or double‐residue substitutions in the IGF loop. (C) Association rate constants determined by BLI experiments using 1 μM ClpP and 2 mM ATP. (D) Dissociation half lives determined by BLI experiments in the presence of 2 mM ATP. In Panels C and D, single‐chain ClpXΔN variants had the IGF‐loop deleted from Subunit B and had a wild‐type or mutant IGF motif in Subunit A. In all panels, values are averages (N = 3) ± SD.
