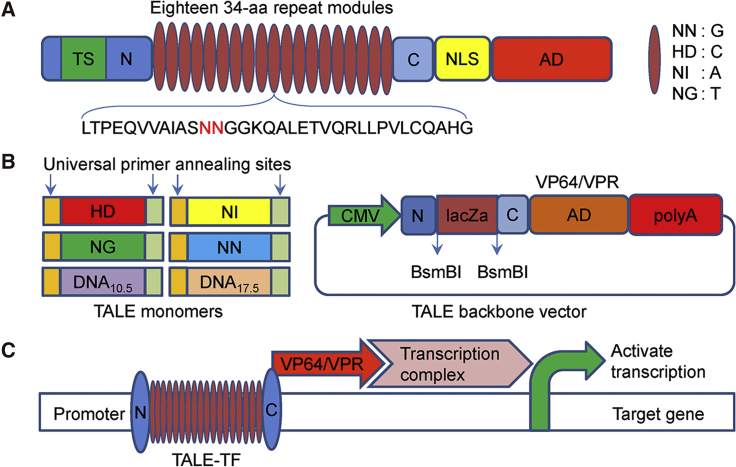
Figure 2.
Schematics of TALE Structure, Monomers, and TALE Backbone Vector for Assembling Custom TALEs
(A) Natural structure of TALEs derived from Xanthomonas sp. Each DNA binding module consists of 34 amino acids, where the repeat variable diresidues (RVDs) in the 12th and 13th amino acid positions of each repeat specify the DNA bases being targeted according to the cipher NG = T, HD = C, NI = A, and NN = G. (B) Monomers and TALE backbones for constructing custom TALEs. Each monomer ends with two constant sequences that provide the annealing sites of a pair of universal primers that can be used to regenerate the monomer library. CMV, cytomegalovirus promoter; N, non-repetitive N-terminus of TALE; C, non-repetitive C-terminus of TALE; BsmBI, type II restriction sites used for the insertion of a custom TALE DNA binding domain; LacZa, LacZa expression cassette for blue-white spot screening; NLS, nuclear localization signal; VP64, synthetic transcriptional activator derived from VP16 protein of herpes simplex virus; VPR, fusion of VP64, p65/RelA, and Rta. (C) Schematic of activating gene expression with TALEs. TALE-TF, TALE transcription factor.