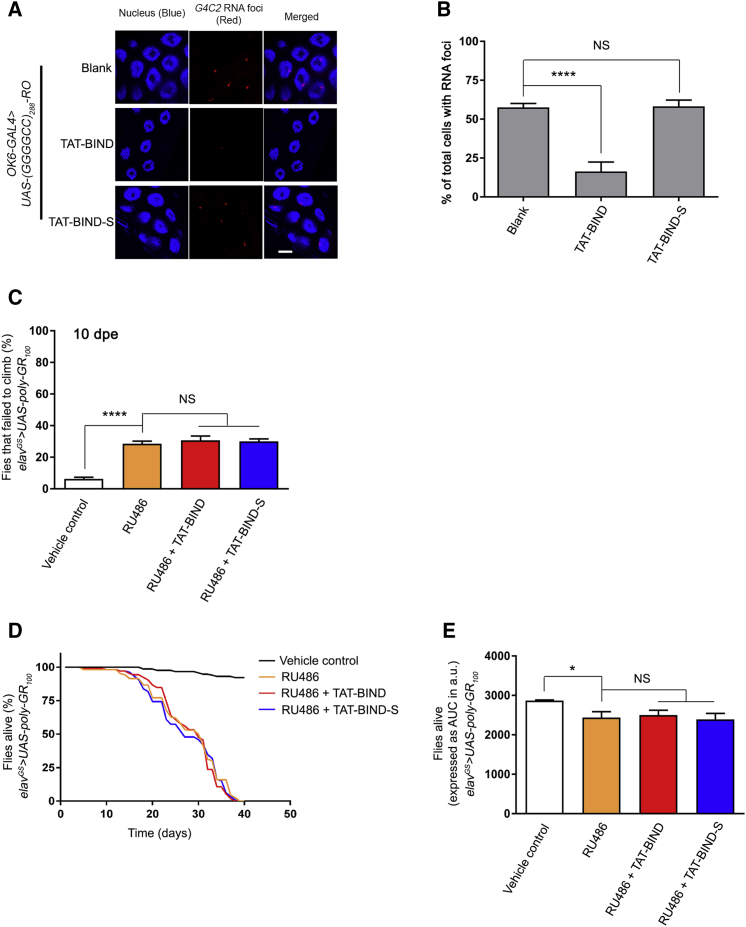
Figure 5.
TAT-BIND Inhibited Expanded GGGGCC-Associated Neurodegeneration in a Repeat RNA-Dependent Manner
(A) TAT-BIND suppressed GGGGCC RNA foci formation in UAS-(GGGGCC)288-RO flies. In situ hybridization was performed to detect GGGGCC RNA foci (red) by using a TYE563-labeled LNA probe. Nuclei (blue) were stained by Hoechst 33332. The scale bar represents 10 μm. (B) Quantification of the percentage of salivary gland cells containing nuclear RNA foci. An OK6-GAL4 driver was used to express 288 RNA-only (RO) GGGGCC repeats in Drosophila salivary glands at third instar larval stage. The genotype was w; OK6-GAL4-GFP/UAS-(GGGGCC)288-RO. At least 4 salivary glands with a total over 100 nuclei were calculated. (C) TAT-BIND treatment did not rescue the climbing defect of UAS-poly-GR100 flies at 15 dpe. (D) TAT-BIND treatment did not prolong the lifespan of UAS-poly-GR100 flies. (E) Area under curve (AUC) analysis of (D). For the climbing ability and lifespan assays, flies at 2 dpe were fed with food containing different drug combinations, including vehicle control (ethanol), mifepristone (RU486, 200 μM), RU486 (200 μM) plus TAT-BIND (50 μM), and RU486 (200 μM) plus TAT-BIND-S (50 μM). Transgene expression was induced with mifepristone (RU486, 200 μM). The climbing ability assay was repeated six times, and at least 90 flies per treatment were scored. The lifespan assay was repeated at least six times, and more than 100 flies per treatment were recorded. The genotype was w; UAS-poly-GR100/+; elavGS-GAL4/+. Data were expressed as mean ± SEM. *p < 0.05 and ****p < 0.0001; NS, no significance.