Figure 1.
Outline of PRISMA Screen and Comparison of Data
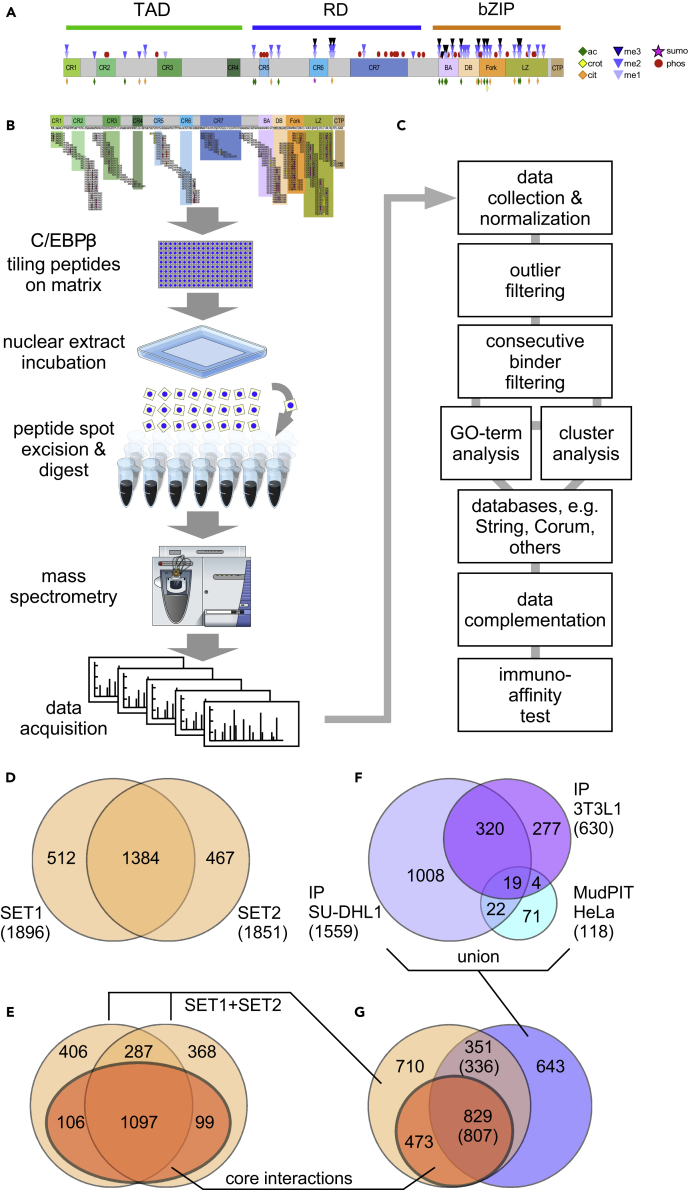
(A) Schematic representation of C/EBPβ and the distribution of known post-translational modifications. Conserved regions (CRs) are depicted in color, whereas intrinsic disordered regions are shown in gray.
(B) Schematic description of the workflow of protein binding and data acquisition.
(C) Workflow of data and proteomic analysis.
(D) Overlap between the two replicates (SET1, SET2) of the PRISMA screen.
(E) Number of proteins in the two PRISMA datasets that show consecutive peptide binding (core interactions, dark amber).
(F) Overlap of three affinity-purification-based datasets of C/EBPβ interactors as described in the literature and combined with data obtained from a proteomic interaction screen in SU-DHL1 cells. Overlaps were determined using immunoprecitation (IP) SU-DHL1 as reference dataset and thus the numbers add up to the size of this set only (see Methods).
(G) Overlap of the PRISMA-derived C/EBPβ interactor datasets from (E) (union of SET1 and SET2, light amber) with core interactions (dark amber) from the union of the three datasets from (F) (blue). Overlaps are given using the PRISMA-derived data as reference datasets. The overlap count using the union of the datasets from (F) as a reference dataset is denoted in brackets.