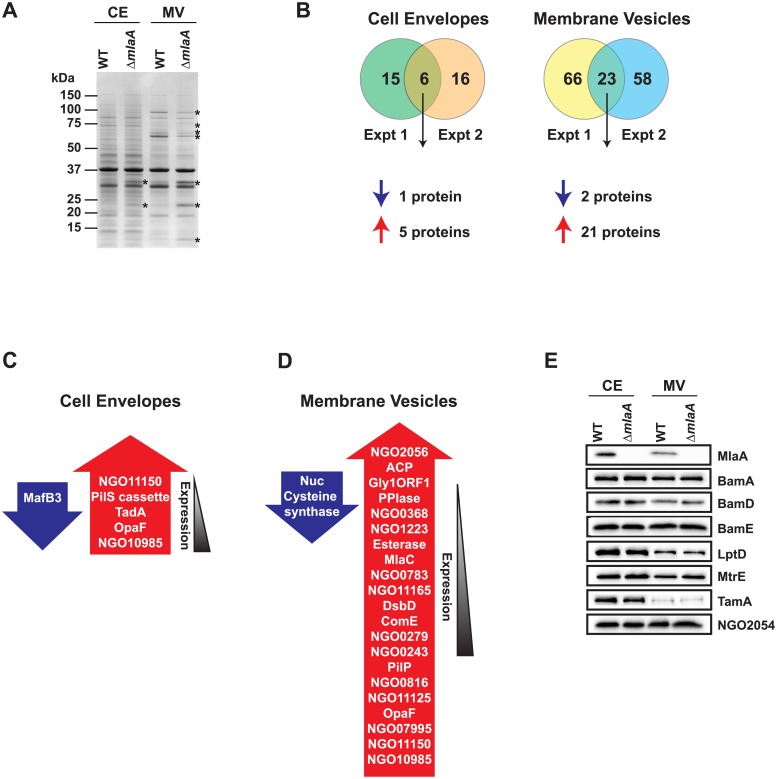
Fig 9. Proteomic investigations of MlaA influence on cell envelope and membrane vesicles.
(A) CE and MV fractions isolated from WT FA1090 and isogenic knockout ΔmlaA were normalized based on protein concentration, separated by SDS-PAGE, and proteins were visualized by coomassie staining. The migration of a molecular weight marker is shown on the left in kDa. Proteins that appeared differentially abundant in the ΔmlaA mutant by visual inspection are labeled with an asterisk. (B) Trypsinized CE and MV proteins from WT and ΔmlaA were labeled with TMT6plex isobaric mass tags, fractionated by strong cation exchange and reverse phase chromatography, and subjected to peptide identification by tandem mass spectrometry. The number of differentially abundant proteins in the ΔmlaA CE or MV protein profiles is noted in the Venn diagrams. (C, D) Lists of differentially abundant proteins in the CE (C) or MVs (D) of the ΔmlaA mutant. Proteins in blue arrows are downregulated in the mutant, while those listed in red arrows are upregulated in the mutant. Proteins are arranged by the magnitude of the mutant:WT ratio. (E) Validation of quantitative proteomics results. CE and MV fractions from WT FA1090 and ΔmlaA were normalized by protein concentration, separated by SDS-PAGE, and probed with antisera against indicated proteins. SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TMT, tandem mass tag.