Figure 1.
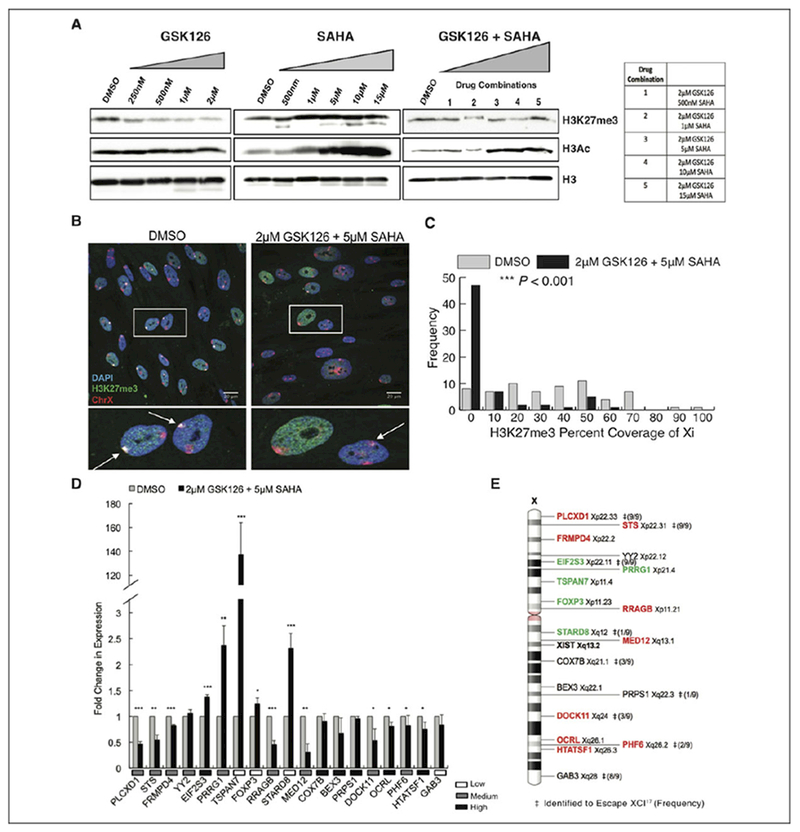
GSK126 and SAHA disrupt chromosome X gene dosage compensation in human female cells. A, Total H3K27me3, H3 acetylation, and histone H3 protein levels of IMR-90s treated with various concentrations and combinations of epigenetic inhibitors, GSK126 and SAHA, or DMSO alone as the control. Total histone H3 was used as a loading control. B, Maximum intensity projections of H3K27me3 (green) and chromosome X (red) localization in IMR-90 nuclei (blue). Insets show enlarged nuclei, arrows highlight location of the Xi. C, Distribution display of 3-dimensional volumetric measurements of H3K27me3 signal over total Xi signal for either DMSO or 2 μM GSK126 + 5 μM SAHA treatment (n = 130 nuclei, 65 nuclei per condition). *P < .05, **P < .01, ***P < .001, Student t test. D, Quantitative PCR analysis of X-linked genes with either DMSO treatment or 2 μM GSK126 + 5 μM SAHA treatment. Values presented as mean (standard deviation) of 3 technical replicates. *P < .05, **P < .01, ***P < .001, Student t test. Relative expression levels of genes in DMSO-treated cells indicated under graph. E, Schematic diagram of chromosome X, with corresponding positions of X-linked genes analyzed in (D) as well as the Xist gene locus. Significantly upregulated genes (green) and significantly downregulated genes (red). ‡Genes identified to escape XCI in human hybrid fibroblasts,17 and frequency of escape listed in parenthesis. DMSO, dimethyl sulfoxide; PCR, polymerase chain reaction; SAHA, suberoylanilide hydroxamic acid; XCI, X-chromosome inactivation; Xi, inactive X.
