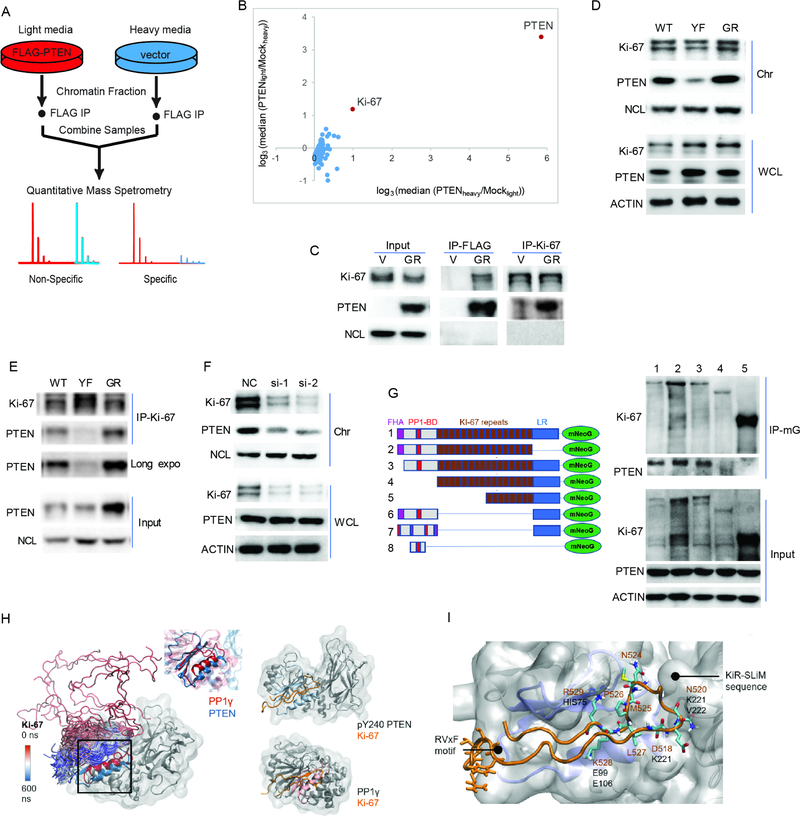
Figure 5. pY240-PTEN binds to chromatin through interaction with Ki-67.
(A) Schematic for the identification of PTEN binding proteins. U87 cells expressing empty vector or Flag-GR-PTEN were labeled in SILAC media containing either light or heavy lysine and arginine. Chromatin fractionation followed by anti-Flag immunoprecipitation (IP) was performed for each sample, and immunoprecipitated proteins were combined and analyzed by mass spectrometry (MS). (B) Scatter plot of the median ratio of PTEN/mock for every protein identified with at least 3 unique peptides from two biological samples were plotted on a log3 scale. Each data point represents a single protein that was identified in this experiment. Data represent two biological repeats by labeling cells with different isotope (heavy or light). (C) Immunoprecipitation of Ki-67 or Flag (PTEN) from chromatin fractions of U87-V or GR-PTEN cells followed by blotting with Ki-67- or PTEN- specific antibody. (D) Chromatin bound Ki-67 in U87-WT/YF/GR-PTEN cells was determined by western blot. (E) Chromatin fractions from indicated cells were prepared for immunoprecipitation with anti-Ki-67 antibody followed by western blotting with PTEN antibody. (F) U87-GR-PTEN cells were transfected with two siRNAs targeting Ki-67 (si-1 and si-2) and a non-specific control (NC). Chromatin fractions were isolated to examine Ki-67 and PTEN levels by western blot. (G) 293T cells were co-transfected with PTEN and Ki-67 truncations (mNeoGreen-tagged). Whole cell lysates were prepared for co-IP with mNeoGreen (mG) antibody followed by western blotting with PTEN antibody; lane 1, Full length Ki-67; lane 2, LR domain lacking (deletion of 2929–3256 aa); lane 3, FHA domain lacking (deletion of 1–134 aa); lane 4, FHA and PP1-BD (deletion of 1–1002 aa); FHA, lane 5, PP1-BD and 8 repeats (deletion of 1–1970 aa). #6, 16 repeats deletion; #7, disrupted FHA and PP1-BD from #6; #8, PP1-BD only. (H) Left: snapshots of Ki-67 trajectory color-coded by the simulation, with the aligned binding pockets of PTEN (blue) and PP1γ (red) highlighted in a square; Center: zoom in at the alignment of the structurally conserved region containing the RVxF and KiR-SLiM binding pockets in pY240-PTEN (blue) and PP1γ (red); Right: comparison between the Ki-67/pY240-PTEN complex formed in simulation (above) and the Ki-67/PP1γ crystallographic complex (below). (I) Close-up to the Ki-67 interaction (orange) with pY240-PTEN, showing the conserved phosphatase fold in blue. The main RVxF motif and KiR-SLiM Ki-67 residues appear in licorice, with the closest interactions labeled. See also Figure S4, Table S5 and movie S1.