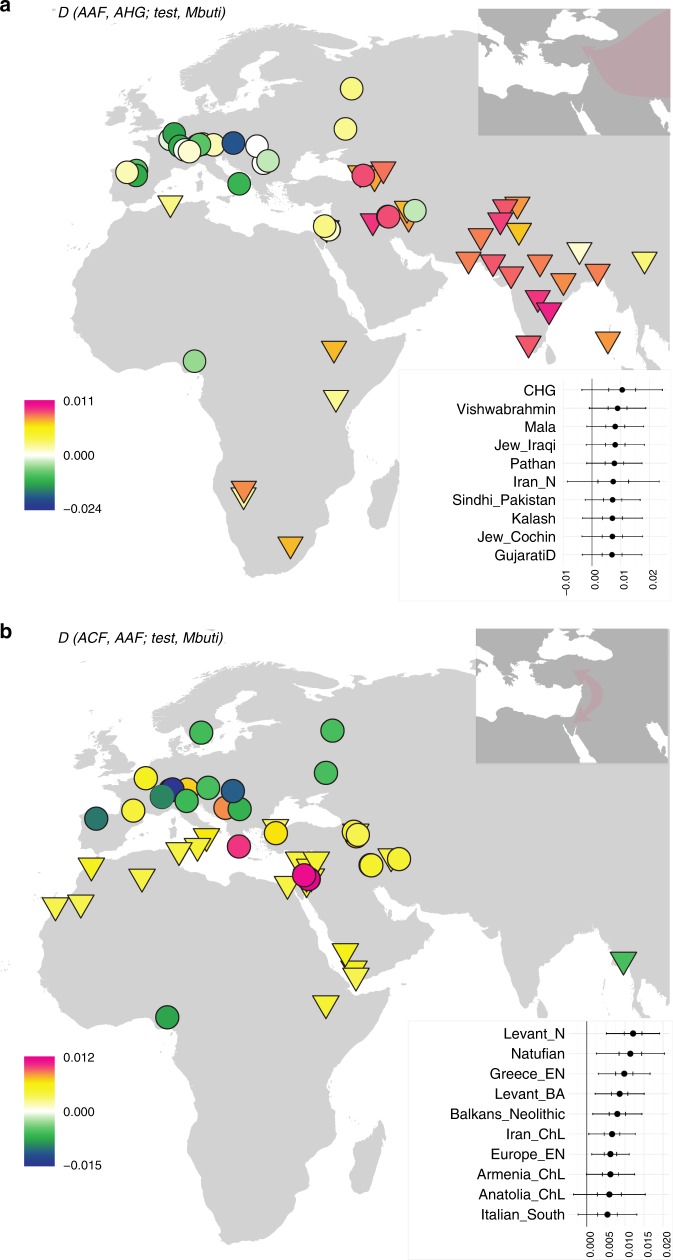
Fig. 2.
Differences in genetic affinities between the ancient Anatolian populations. We plot the highest and lowest 40 values of D(population 1, population 2; test, Mbuti) on the map. Circles mark ancient populations and triangles present-day ones. “Test” share more alleles with population 1 when values are positive and with population 2 when negative. The detected gene flow direction is illustrated in the upper schematics; the illustrated rout represents the shortest one between the proximate source and the target and should not be interpreted as the historic rout of the gene flow. The statistics and SEs are found in Supplementary Figures 2–5 and Supplementary Data 3. a Early Holocene Iranian and Caucasus populations, as well as present-day South Asians, share more alleles with Aceramic Anatolian farmers (AAF) than with Anatolian hunter-gatherers (AHG), measured by positive D(AAF, AHG; test, mbuti). The top 10 values with ±1 and ±3SE are shown in the upper box. b Ancient Levantine populations share more alleles with Anatolian Ceramic farmers (ACF) than with AAF, measured by positive D(ACF, AAF; test, Mbuti). The top 10 values with ±1 and ±3 SE are shown in the lower box. Source data are provided as a Source Data file