Short abstract
Genomic instability has been an area of active area of research in the last two decades. Based on the initial study for hereditary cancers, DNA repair gene family mutations have been identified. In sporadic (non-hereditary) cancers, several large-scale DNA sequencing studies suggest that mutations in DNA repair genes are less frequent, suggesting the complexity of tumorigenesis of sporadic cancers. So far, several important genes have been identified, by using mostly cell line models and mice studies. These include DNA damage response modifier like ataxia telangiectasia mutated (ATM), conventional tumor suppressor genes like TP53 and cyclin-dependent kinase inhibitor 2A (CDKN2A; which encodes p16INK4A and p14ARF). Various hypotheses for sporadic tumorigenesis have been developed, and so far, “oncogene-induced DNA replication stress model” has been gaining widespread interests. In this review, we will first describe some of the important concepts of genomic instability. Then, we will outline oncogene-induced genomic instability and discuss the role of the MYC gene during this process, which will be followed by detailed reviews of mutation data. We hope that this review can outline the overall perspectives of genomic instability.
Impact statement
This review provides various genetic and cell line data previously published in a way to explain how cellular stress can lead into genetic instability.
Keywords: Gene mutations, cellular stress, oncogene, tumor suppressor gene, p53, MYC
Introduction
Eighteen years ago, “six functional characteristics common to human cancer”, described as “hallmarks of cancer”, were proposed to describe the common biological characteristics of cancer cells.1 These six hallmarks were well understood from several decades of research, while the molecular mechanistic of such hallmarks have not been fully explained yet. These six hallmarks include following biological nature of cancer cells: constant proliferative signaling, overcoming growth suppressors, cell death resistances, immortalized cell replication, angiogenesis promotion, and increased invasion and metastasis potential. While these concepts have been continuously updated to provide a better understanding of the molecular basis of human carcinogenesis, recent progress in the cancer biology is still in its early stage to fully dissect molecular mechanism behind. Of note, accumulated knowledges from various DNA sequencing studies using cancer samples have taught us that DNA mutations do not seem to be acquired in an orderly fashion. A new concept of “genomic instability” was developed to provide a mechanistic model for cancer cells for their ability to develop and maintain part or all of these six hallmarks.1
While genomic instability is a fundamental process of almost all human cancers, the exact role of these comprehensive processes in each stage of tumorigenesis is still unclear. Chromosomal instability (CIN), a term describing changes in chromosome structure and number, happens over a long period of time and certainly happens during normal cellular life spans. However, the frequency and magnitude of CIN are way more profound in cancer cells as compared with normal cells. For a long time, CIN has been visualized,2,3 and it has been widely accepted theory that a genetically unstable single cell can result in multiple progenies of tumor cells by continuously acquiring chromosomal abnormalities.4,5 While CIN develops over a period of long time for sporadic cancers, a defined set of genomic instabilities can happen for certain kinds of cancer cells, which can explain some of hereditary tumors. These include microsatellite instability (MSI), a process in which oligonucleotide repeats present in certain DNA sequences fluctuates,6,7 and other forms of genomic instability that are derived from a lack of proper DNA repair system, which results in increased frequencies of base-pair changes.8
Although the hallmarks of cancer are hard to group into several simple categories, it is generally believed that the interaction between tumor cells and the immune surveillance system plays a crucial role in tumorigenesis, and therefore evading the immune surveillance9 should be an important component. In addition, several unique defense mechanisms against cellular stresses for tumor cell survival will be important. Thus, recent cancer researchers have focused on how cancer cells cope with these various levels of cellular stresses, and therefore five additional hallmarks are proposed: oxidative stress, proteotoxic stress, metabolic stress, DNA damage and DNA replication, mitotic stress.10,11
Overall, while these hallmarks are different from each other in their mechanism of development and biological impact to the cancer cells, they all are significantly affected by genomic instability, which is not only a driving force of initial tumorigenesis but also for late evolution of human carcinogenesis. Therefore, in this review, we present prior understandings for genomic instability, a review of large-scale DNA sequencing, a new theory of oncogene-induced tumorigenesis and a detailed review of mutations in various cancers as related with genomic instability.
Overview of genomic instability
Hereditary versus sporadic cancers
In hereditary cancers, mutations found within DNA repair genes have been studied as one of the key drivers of genomic instability. So far, there have been several exciting findings from a careful analysis of pedigrees gathered from high-risk cancer-prone family members. Overall, discovery of novel mutations in DNA repair genes in several hereditary cancers has fermented the idea for “the mutator hypothesis.” This hypothesis suggests that genomic instability present in precancerous cells acts as a driving force for tumor development by gradually elevating the rate of spontaneous mutation.4,12 In fact, for various inherited cancers, germ-line mutations in DNA repair genes are often accompanied by secondary loss of the wild-type gene copy. This loss of remaining “good copy of gene” seems to be a mandatory step for tumor development. However, in most sporadic cases, the mutation profiles are far more complicated.
One of the first examples supporting this “mutator hypothesis” was the discovery of a set of genes underlying hereditary non-polyposis colon cancer, where several mutations in genes encoding DNA mismatch repair enzymes cause MSI.6,7 Likewise, in hereditary MYH-associated polyposis, DNA base excision repair (BER) gene mutations are discovered, and this BER gene mutations seems to result in high-frequency base changes from G•C to T•A.8 In more common hereditary cancer patients, mostly breast and ovarian cancer, which are accompanied by the presence of significant CIN, a novel mechanism of repair pathways has been characterized. These germ-line mutations are best seen in two related genes: breast cancer susceptibility 1 and 2 (BRCA1 and BRCA2).
Subsequently, several partner proteins of BRCA1 and BRCA2 were discovered and characterized. These proteins often colocalize with either of BRCA1 and BRCA2 and expanded our understanding of various DNA repair processes and DNA damage responses (DDRs). The scope of molecular interactions between BRCA1/2 and these partner proteins are beyond our review. However, it is worth to note that these partner proteins are rather extensive: PALB2, BRCA1-interacting protein 1, Bloom syndrome helicase, RecQ protein-like 4, the Fanconi anemia genes RAD50, Nijmegen breakage syndrome protein 1,13 Werner syndrome helicase. Expectedly, mutations of these partner genes have been also linked to the development of various cancers, including not only in breast and ovarian cancer but also leukemia, lymphomas,14–16 and skin cancer.13
Mutations of caretaker genes and oncogene-induced DNA replication stress models
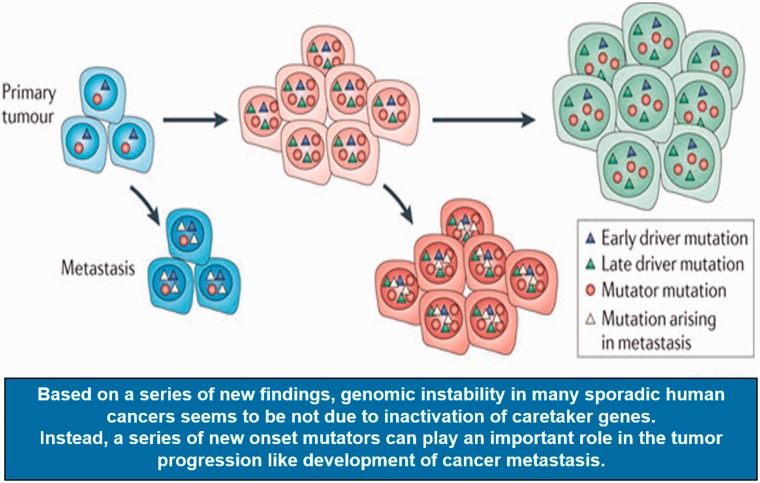
As described above, germ-line mutations in caretaker genes can provide logical explanations for the presence of genomic instability in various inherited cancers. The DNA repair genes and mitotic checkpoint genes are two classical caretaker gene family, and most of the time, these two classes of gene family do not undergo inactivation during carcinogenesis. In contrast, non-classic-caretaker genes like the tumor suppressor gene TP53 and the ataxia telangiectasia mutated (ATM), in which both genes are crucial in the DDR, are under selective pressure for inactivation during tumorigenesis and lose their functions in significant portion of cancer cells. While several efforts to identify “classic caretaker genes” in sporadic cancers have been attempted, the discovery of such genes as a “major inducer of human cancers” has been largely disappointing.17 For example, an initial DNA sequencing analysis of 100 DNA repair and cell cycle checkpoint genes in human colon cancer cell lines, in their early stage of passages, identified only very few mutations.18 Likewise, DNA sequencing analysis for the mitotic checkpoint gene budding uninhibited by benzimidazole 1 (BUB1 gene) rarely identified any meaningful mutations in human cancer samples, while in experimental models, BUB1 gene can cause significantly elevated CIN.19,20 Based on the above results and various other reports,11 it is now widely accepted that genomic instability in many sporadic human cancers is not due to the inactivation of caretaker genes. In fact, so far, various DNA mutation studies in multiple sporadic cancers suggested that more than 69% of cancers did not harbor mutations in caretaker genes. Instead, it seems that a series of new onset mutators, which can happen randomly unlike genes mutated in hereditary cancer, may play an important role in the tumor progression like initiation of tumorigenesis to the development of cancer metastasis (Figure 1).
Figure 1.
Accumulated mutations can promote tumor metastasis. Metastatic spread may occur throughout tumor progression, both early in tumor evolution and as the tumor evolves more genetic alterations. The metastases may then evolve to develop metastasis-specific mutations that are different to those found in the primary tumor (modified from Loeb12).
Thus, unlike hereditary cancers, which are driven by few caretaker gene mutations under the “mutator hypothesis,” the genomic instability in sporadic cancers seems to be difficult to be explained by “mutator hypothesis.” Likewise, even in the same organ origin like in the case of lung cancer, depending on type of histological findings, different type of cancer clearly carries not only different type of mutations but also has shown that such mutation frequency varies. In fact, through analysis using cell lines carrying various transfected oncogens, the oncogene-induced DNA replication stress model has been established and now is widely accepted as a leading hypothesis for sporadic cancer development.10,21–24 For example, CIN in sporadic cancers can be explained from the observation that the transfected oncogenes into normal cells can induce DNA replication forks collapse with accompanying catastrophe in DNA replications, DNA double-strand breaks (DSBs) and accelerated CIN, all of which are hallmarks of genomic instability.
Recent efforts for high-throughput sequencing studies
In efforts to identify novel mutations in primary cancers, cancer cell lines in their early passages and tumor xenografts, several consortia have been formed. In one of the first studies done by Vogelstein and his collaborators, 18,191 genes have been sequenced from early passage xenografts or cancer cell lines from a limited 11 breast and 11 colorectal cancers.25,26 This study demonstrates a systemic way of analyzing gene mutations, starting from cell lines into cancer tissue samples. At first, in the cell lines from breast cancers, various mutations were identified in 1137 genes (initial discovery screen). Based on this, an additional 24 breast cancer tissue samples were analyzed, and mutations in 167 genes were discovered (validation screen). Likewise, in the colorectal cancers, from the initial discovery screen, 848 genes with mutations were identified. In the validation screen, mutations were found in 183 of the 848 genes from 24 additional colon cancer tissue samples.
As a follow-on study, the same group of investigators analyzed the coding sequences of more than 20,000 genes from 24 advanced pancreatic adenocarcinomas27 and glioblastomas.22,28 In this study, more extended number of samples were analyzed for genomic deletions and amplifications by single nucleotide polymorphism arrays. Overall, the results showed a similar trend to the prior reports. From the initial discovery screen, in the case of pancreatic cancer, more than 1300 genes (1327 genes) were mutated in at least one sample. In the case of glioblastomas, more than 650 genes (685 genes) were mutated at least one sample. In subsequent validation phase, 39 genes were sequenced in an additional 90 pancreatic cancers, and 21 genes were sequenced in an additional 83 glioblastomas.
The largest number of cancer cases were examined from lung cancer samples by several groups from the Cancer Genome Atlas Research Network, and a total of 623 cancer-relevant genes were sequenced in 188 primary lung adenocarcinomas.29 Out of 623 cancer-related genes, more than half of genes (356 genes) were mutated at least once. Of note, 193 of 356 genes were mutated more than one sample, while various genomic changes including gene amplifications and deletions were also discovered. Additionally, The Cancer Genome Atlas Research Network reported similar kinds of analyses for glioblastoma samples, which included both untreated and treated patients.30 In this report, 601 cancer-relevant genes were analyzed from biopsy or resected tumor tissues of patients with untreated (72 cases) and treated glioblastomas (19 cases). In a total of 91 cases including the combined treated and untreated cases, 223 genes were mutated, while 79 of 223 were mutated in more than one sample.
Unexpected findings from high-throughput sequencing studies
By surprise, while many genes were mutated from all of the large-scale DNA sequencing studies as discussed above, only a few genes harbored point mutations, amplifications, or deletions with reasonable high frequencies.25–30 For example, in each cancer types analyzed, only about four genes were altered in more than 20% of the tumor samples and that three most commonly altered genes are TP53, epidermal growth factor receptor (EGFR) and the small GTPase RAS, in addition to other less commonly mutated genes. It is therefore important to understand the function of these frequently mutated genes and their gene families.
Foremost, the TP53, which plays a key role as a tumor suppressor and a major mediator of DNA damage checkpoint, was mutated most frequently from all tumor types, while several oncogene products were among the frequently deregulated genes including EGFR and the small GTPase RAS. In addition, other tumor suppressor products stand out, although not as frequently as TP53. These include neurofibromatosis type 1 (NF1), the phosphatase, tensin homologue deleted on chromosome 10 (PTEN), and the cyclin-dependent kinase 4 inhibitor p16INK4A. As expected, based on different tumor biology, frequently deregulated growth-regulating genes seem to be different among different tumor types. For example, in pancreatic and lung adenocarcinomas, RAS mutations were frequent but were essentially absent in glioblastomas. However, in the case of glioblastomas, mutations from three genes, NF1, PTEN and EGFR, were the most prevalent, while RAS mutations are rare. While these findings are rather unexpected, a follow-on in vivo mice study has been designed based on these tumor-specific mutation profiles.
Oncogenes induce genomic instability
While many genes were mutated from all of the large-scale DNA sequencing studies as discussed above, caretaker genes mutations so far has not been able to explain genomic instability observed in many sporadic human cancers,25–31 and therefore genomic instability is suggested to originate from mutations in other genes. While genomic instability like CIN are hallmarks for almost all sporadic human cancers,2,3,5 it had been still difficult to define molecular mechanistic behind human carcinogenesis. Instead, it has been long hypothesized that the genes responsible for genomic instability in sporadic cancers are likely to be genes mostly mutated in human cancer samples.
Therefore, it has been originally thought that oncogenes are unlikely group of genes causing genomic instability,32 while inactivation of TP53 as a DNA damage checkpoint gene33 was expected to cause genomic instability. However, cell line studies, mice model, and human tissue analysis did not support this idea. Foremost, TP53 gene deletions in mouse models and human cells did not cause aneuploidy, which is one of the most common forms of CIN.34,35 Second, from the careful analysis of various human precancerous tissue samples, genomic instability was evident even before TP53 mutations were firmly established.21,24 These two observations in addition to the lack of mutations in caretaker genes have resulted in conceptual changes for the role of oncogenes in inducing genomic instabilities. Therefore, by using various cell line models, the role of oncogene-induced genomic instability has been actively pursued. Surprisingly, the activation of oncogenes in cultured mammalian cells, by oncogenic RAS or MYC which can induce a strong growth signaling, resulted in the key features of genomic instability including accelerated CIN and loss of heterozygosity. Subsequent in vivo studies using human cancer cell xenografts and various mouse models replicated findings from cell line studies.21–24,36–43
While exact molecular pathways need to be clarified, it seems that the prevailing theory can be summarized as “oncogene-induced DNA replication stress model.”10 This model is based on the hypothesis that activated oncogenes induce genomic instability by causing DNA replication stress and associated DSBs. DNA replication stress happens when cell replication is significantly promoted during tumorigenesis, resulting in mutations in specific genomic sites, which was described as “common fragile sites.”44 In fact, unlike for the case of TP53, in human precancerous lesions and cell lines systems where oncogenes have been activated, genomic instability is not only induced but also seems to preferentially affects common fragile sites.21–24,45
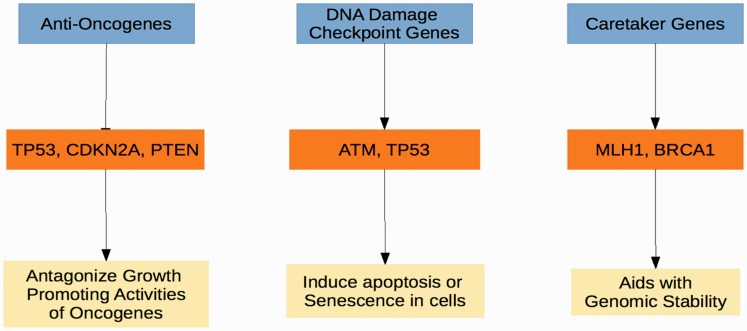
Then, how can these oncogenes-induced replication errors be linked to the frequent mutations found in tumor suppressors like TP53, CDKN2A or even PTEN? These questions are important in a sense to reconcile between the old beliefs that mutation in tumor suppressors can induce genomic instability and several recent observations of oncogene-induced genomic instability. This is a significantly complex process, and the observations we have are likely just the tip of the iceberg. However, we can summarize three distinct types of tumor suppressors and their relationship during oncogene-induced genome instability (Figure 2). Here we discuss some of the most recent concept of oncogene-induced genomic instability.
Figure 2.
Tumor suppressor gene classes. Three well-defined tumor suppressor classes can be defined based on the primary function of the proteins they encode. Anti-oncogenes function by encoding proteins that antagonize oncogene activity, such as with CDK4 (CDK4) and cyclin D1 (CCND1) being inhibited by the protein products of cyclin-dependent kinase inhibitor 2A (CDKN2A) and retinoblastoma (RB1). DNA damage checkpoint genes respond to DNA damage and replication stress by inducing cell death or senescence via ataxia telangiectasia mutated (ATM) and TP53. Caretaker genes such as MLH1 and BRCA1 promote genomic stability.
First, oncogene-induced DNA damage rapidly activates TP53 as a key player of DNA stress response. If TP53 fails to repair damaged DNA, it can then activate TP53-dependent death pathways, mostly cellular senescence followed by inevitable apoptosis. Therefore, activation of TP53 can limit the growth of the pre-neoplastic cells and precancerous lesions surrounding tumor mass.
However, when the function of TP53, based on accumulated mutations, is lost as a secondary response to oncogene-induced DNA damage, cells can escape its apoptotic and/or senescence effects. This series of event, from oncogenic activation to the loss of TP53 function, may be the point when the precancerous lesion can then become fully cancerous.21–24 Second, while the product of TP53 which, among other numerous functions, plays a pivotal role as a DNA damage checkpoint protein responding to oncogene-induced DNA damage, most other tumor suppressors compromise the same growth signaling pathways as inhibitors of defined sets of oncogenes. For example, the PTEN tumor suppressor gene product (PTEN phosphatase) antagonizes the function of phosphoinositide 3-kinase-α, one of key oncogenic kinase.46 Likewise, as a CDKN2A tumor suppressor gene product, the p16INK4A protein directly antagonizes the cell cycle-promoting activities of two crucial kinases (CDK4 and CDK6 kinase) encoded by the two related oncogenes, CDK4 and CDK6.47
Third, in certain cases, alternative route linking tumor suppressor dysfunction with oncogene-induced DNA damage has been reported. Most recently, chronic expression of the tumor-suppressor p21WAF/Cip1, in a p53-deficient environment, was shown to exhibit an oncogenic behavior.48 This led to “bypass” of the antitumor barrier of senescence, resulting in the emergence of “escaped” clones that demonstrated genomic instability, increased aggressiveness, and chemo-resistant features. Unexpectedly, sustained p21 accumulation was shown to cause deregulated origin licensing of DNA replications accompanying replication stress. It seems that this surprising effect of p21 is secondary to the inhibition of the CRL4–CDT2 ubiquitin ligase. These observations clearly suggested that a novel tumor-promoting activity of p21 can be secondary to its ability to deregulate DNA replication, particularly replication licensing. Of note, it has been well established that p21 responds to various genotoxic stress like exposure to chemotherapeutic agents or radiation; thus, this induced p21 activity has been suggested to be one of the key mediators of the response from various cancer therapeutics.11 Fourth, the caretaker genes,4,12 most of which are DNA repair genes, are a group of genes that still can be considered as a unique class of tumor suppressor gene. While these gene families play a crucial role in maintaining genomic instability, in response to genotoxic stimuli, they seem not to directly affect cell growth or senescence with follow on apoptosis.
MYC-induced replication stress and genomic instability
The c-MYC (MYC) is one of the most frequently altered genes in cancer and one of the first identified human oncogenes. Basically, MYC, as a proto-oncogene, controls a wide process of cellular biology including cell growth, cell death, cell cycle progression, and energy metabolisms by governing expression of a very large number of target genes.49–51
The cellular functions of MYC is under the tight controls not only through multiple transcriptional but also post-transcriptional regulatory mechanisms, as ensuring a precise control of MYC protein levels in proliferating cells are crucial for cellular survivals. Therefore, deregulated MYC expression secondary to different types of genetic mutations or even epigenetic changes of its promoters can result in constitutive activity. In fact, in different cell types, multiple cancer cell lines, and in transgenic mice models,52–55 it has been demonstrated that MYC is constitutively activated and therefore promotes oncogenesis. The mechanism whereby MYC contributes to tumorigenesis seems to be of more than one route. It can cause overstimulation of cell growth and also can make drastic changes in cellular metabolism, both of which then contribute genomic instability.56 Both of these effects seems to confer MYC to induce genomic instability by causing accelerated DNA damage with increased mutations, enhanced gross chromosomal rearrangements, both of which then can induce inappropriate cell cycle progression,57 eventually promoting tumorigenesis.
Fundamentally, MYC can control cell proliferation and cause genomic instability in both normal and tumor cells. Initially, it seems clear that at least part of this is secondary to MYC’s ability to regulate DNA replication based on both transcriptional and non-transcriptional mechanisms. Mechanistically, DNA replication is a tightly controlled process and initiation of DNA replication heavily depends on three major steps, all of which are finely regulated. These include initial assembly of the pre-replicative complex at the origin of DNA replication (late mitosis/early G1), activation of these sites (transition of G1 to S),58 and accompanying highly coordinated epigenetic modifications including remodeling of nearby chromatin. Any changes in these time- and space-controlled processes can lead into significant changes in cell cycle and cellular fate. From the aspects of MYC, the subjects of research has been highlighted on “How uncontrolled MYC expression can compromise each of these three steps of coordinated DNA replications?,” as we now understand that compromise of any of these three steps can result in unscheduled DNA synthesis, dysregulated checkpoint activation, and genomic instability.58 As MYC controls expression of key genes for cell cycle, MYC was initially proposed as one of the masterminds regulating cell proliferation. It has been subsequently shown that as discussed below in detail, MYC can directly control activity at the DNA replication origin, which is unexpected but clearly based on the non-transcriptional mechanism.23
How proper DNA replications are protected?
In order to understand the concept of MYC-induced genomic instability, it is important to understand molecular mechanisms, based on which cells can be protected from accumulated DNA damages and dysregulated cell proliferation. Initiation of DNA replication in eukaryotic cells starts from multiple replication origins located in each chromosome. While this process is complicated and certainly is most energy efficient way for the large eukaryotic DNA to replicate, it can still result in multiple replication errors. Therefore, a tight controlling mechanism exists in two different levels. First, there are carefully designed mechanisms to secure that DNA replication occurs only one time per each cell cycle. Second, when any mistakes are made during DNA replications, it is important to monitor the degree of DNA damage and prevent DNA replication until such DNA damages are completely repaired.54 Cell has its own system to fulfill this function, through a process called “checkpoint responses.” For example, dysregulated MYC expression results in DNA damage, and in such cells with MYC-induced DNA damage, “checkpoint response,” ATM/Chk2 and ATR/Chk1 kinase pathways are activated to immediately stop the cell cycle progression and allow time for cells to repair its own damaged DNA. While details are beyond the scope of this review, both ATM and ATR are upstream regulators of cell cycle checkpoint signals and their function depends on, through their unique roles as upstream kinase, activation of key checkpoint-related proteins including p53, Chk1, and Chk2.23,50
How does c-MYC induce DNA stress response in quiescent cells?
For normal cells differentiated into tumor cells, there should be mechanism where DNA replication must be eventually continued without interference from checkpoint response pathways, which should follow into genomic instability. Due to a known property of c-MYC that has a strong oncogenic property leading to genomic instability, there have been several efforts to link proteins involved in gene expression in quiescent cells and c-MYC. One of the best-documented examples is the interaction between c-MYC, p300, and CBP. These two closely transcriptional factors, p300 and CBP, are well characterized nuclear phosphoproteins, in terms of their function as transcriptional coactivators in various cell line models. Of note, these two proteins are unique in a sense that both of them carry innate enzymatic activity of histone acetyltransferase and can acetylate histone proteins with resulting chromatin remodeling.59 Importantly, suppression of p300 or CBP in resting cells can induce c-MYC, which then induces DNA synthesis even in the deficiency of serum in cell line culture system or lack of growth factor stimulation.60,61 Furthermore, conventional DNA tumor viruses like adenovirus E1A or SV40 large T protein, both of which can cause cellular transformations, also deactivate p300/CBP, through their direct binding activity, thereby inducing c-MYC. Therefore, it seems that these DNA tumor viruses, after infection, can induce S phase in resting cells even without support from external mitogen.60 While, in p300 suppressed cells, deregulated/early onset DNA replication allows them to get out of G1 phase and enter S phase, they then accumulate mostly in S phase and is not able to enter G2/M and ultimately die due to inevitable apoptosis.61 Because serum addition to cells without p300/CBP also results in elevated c-MYC synthesis and a block in S phase, inability of cells without p300/CBP to enter G2/M from S phase seems to be unrelated with status of growth factor signaling.61 In summary, inappropriate/overly stimulated activity of DNA replication origin by c-MYC results in replication stress, S-phase block with lack of G/M phase, all of which cause widespread DDR, suggesting an important guardian role of p300 in maintaining genomic stability.
MYC controls genomic instability by modulating DNA replication origin
How does MYC induce DNA replication stress? This is a challenging question, and it is important to understand how replication starts as oncogene-induced genomic instability is based on DNA replication forks collapsing, which are preceded by abnormal DNA replication origin.
DNA replication involves the sequential assembly of the pre-replicative complex on chromatins near replication origin.58,62,63 The origin recognition complex is recruited first, followed by loading of DNA helicase, the mini chromosome maintenance (MCM) 2–7. This process is mediated by the Cdc6- and Cdt1, and while MCMs are present in ample amount, they are largely inactive double hexamers, and therefore it can be activated only after DNA loading.63–65 Replication initiation requires S phase CDKs and CDC7, which then results in MCMs binding to Cdc45 and GINS complex and activates helicase function of MCMs.63,66 Once DNA is unwound by the Cdc45-MCM2-7-GINS (CMG) complex,67–70 the DNA polymerase is inserted into replication origin to initiate DNA replication.71–73 In contrast to MCMs, that are loaded in excess,74 Cdc45 is limited.75,76 Therefore, Cdc45 is now considered to be one of the useful indicators for replication forks in action.77 As MYC overexpression promotes excessive binding of Cdc45 onto chromatin,78 the activated Cdc45, after binding to chromatin, can result in deregulated initiation of DNA replication. This can be detrimental for genomic integrity, as overly activated Cdc45 can lead late DNA replication origins to start earlier than early origin, which results in the S phase chaos.79
Most recently, Srinivasan et al.80 have provided a more detailed evidences, although similar to previous report as discussed above.78,79 Basically this report demonstrated that Cdc45 is a direct MYC downstream target and mediates DNA replication stress driven by MYC. This is based on the observation that overexpression of MYC and, to a lesser extent, overexpression of GINS result in four different effects on DNA replication and genomic instability, all of which are also driven by MYC. These include an increased early origin firing, a decreased inter-DNA replication origin distance, an increased amount of asymmetrical DNA replication forks, and accumulated DNA damages. This report concludes two important findings. First, it suggests that MYC overexpression enhances Cdc45 recruitment to replication origins. Chromatin-bound Cdc45 then overly activates DNA replication by deregulating activation of replication origin. Deregulated firing of replication origin in turn, results in replication catastrophe including stalling of replication fork, leading to inevitable genomic instability. Second, it did confirm the notion that CDK2/cyclin E complex plays a key role in the activation of replication origin, while MYC regulates origin firing by controlling activity of CDK2/cyclin E complex.
In summary, these reports in addition to many other studies support the idea that Cdc45 and CMG (two critical determinants of the DNA replication origin) complex can partner with MYC overexpression in tumor development, and this makes these two genes as novel oncogenes.
Models of oncogene-induced genomic instability from CML
Human leukemia is characterized by genomic instability, of which chromosomal rearrangements are well documented.31 A well-established way in which DNA rearrangements or mutations can be generated is through the improper induction and repair of DSBs.81 These can be induced in cellular DNA through increased production of reactive oxygen species (ROS).81
Recent studies have shown that several forms of myeloid leukemia, including Philadelphia chromosome 1-positive chronic myeloid leukemia (CML), generate endogenous DSBs with concomitantly increased frequencies of improper repair of this DNA damage through non-homologous end-joining (NHEJ), one of the two pathways for the DNA damage repair pathways of DSBs in mammalian cells.82 These authors elegantly demonstrated that the oncogenic BCR–ABL fusion gene, resulting from the reciprocal translocation of chromosome 9q34 with 22q11 [(t9; 22) (q34;q11)], present in cells of the vast majority of CML patients, produces increased ROS, which in turn gives rise to increased DSBs.
Strikingly, the increased ROS and DSBs (quantitated by assays such as phosphorylated histone variant γH2AX foci formation) are dramatically reduced by agents that inhibit DSB repair, block the BCR-ABL tyrosine kinase activity (STI571), or inhibit generation of ROS (the antioxidant, pyrrolidine dithiocarbamate (PDTC)). The authors found that subsequent repair of endogenous DSBs by both NHEJ and another more proper DNA repair pathway for DSBs, homologous recombination repair (HRR) were also enhanced. However, a significant percentage of these DSBs are repaired incorrectly leading to an increase in the frequency of DNA deletions and mutations. This is a good example for how BCR-ABL, an oncogenic product, can induce genomic instability by improper DNA repair mechanisms signaled by ROS. It is interesting to see that in this case, replication stress is signaled by ROS, which probably causes excessive DSB in turn causes excessive repair signals.
An integrated model: Inactivation of tumor suppressors and activation of oncogenes
The key of the “oncogene-induced DNA replication damage model” starts with a process whereby oncogenes induce both DNA replication stress and DSBs, leading to activation of stress signal molecules like ATM (Figure 3). In normal cells, these stress signals usually either induce DNA repair or cellular death through various pathways including death pathways mediated by p53. However, in cells that become tumorigenic, these two fundamental processes seem to be compromised.
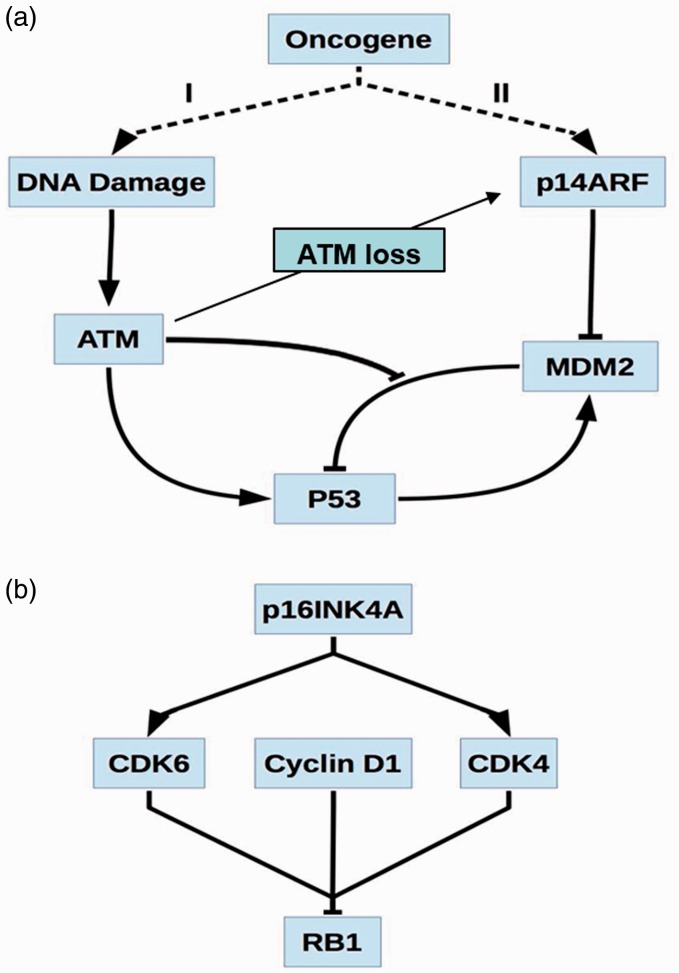
Figure 3.
Proposed DNA replication stress and their corresponding pathways. (a) Two proposed DNA replication stress models induced by oncogenes. Oncogenes of Model I induce double-strand breaks (DSBs) and replication stress leading to the activation of ataxia telangiectasia mutated (ATM). ATM follows by activating p53, while also disrupting the interaction of p53 with MDM2 inhibiting MDM2-dependent p53 breakdown. Model II proposes that p14ARF is activated by oncogene activity, causing MDM2 inhibition and p53 activation. It is interesting to note that loss of ATM signal can stablize ARF signal, and therefore ARF can play as a second line of defence mechasnims. (b) An alternative involving CDKN2A is believed to cause activation of p16INK4A, an inhibitor of cyclin-dependent kinase 4 (CDK4) and CDK6. When CDK4 and CDK6 are combined in a complex with Cyclin D1, they act in concert to inhibit the effects of retinoblastoma 1 (RB1).
In a simple mechanistic model, oncogene-induced ATM activates p53 and modulates activity of p16 and ARF. Any failure of these two proteins by various mechanisms can induce tumorigenesis. The activation of p53 primarily depends on protein stability from disruption of the interaction between p53 and the E3 ubiquitin ligase MDM2, which results in stabilization of the p53 protein. While it was originally suggested that the function of p53 happens mainly within the nucleus, p53 also plays a direct role in inducing apoptosis in cellular cytoplasm. It is important to note that oncogenes-activated p14ARF can inhibit MDM2 function by directly binding to MDM2, which result in reduced interaction between MD2 and p53. This is an important mechanism for oncogene-induced p53 stabilization and activation (Figure 3(a)). In the case of p16INK4A, it can directly inhibit both of two cyclin-dependent kinases, CDK4 and CDK6, and in combination with cyclin D1, it can compromise the tumor suppressor function of the retinoblastoma 1 (RB1) protein (Figure 3(b)).
Recently, two interesting reports have described interesting connections between DDR and induction of ARF, which merits detailed discussion. As discussed above, oncogenic induction in transfected cells results in the DDR and triggers the expression of ARF tumor suppressor, both of which cause activation of the p53 pathway. However, the question remains regarding “why cell needs both pathways?” and “what is a time frame between ARF induction and DDR activation?” Moreover, the strength of stress required for ARF induction and DDR activation is also important question to fully understand “oncogene-induced genomic instability.” These three questions presently become an important topic of cancer research.
More recently, Evangelou et al. have addressed these questions by analyzing mouse models, each carrying colon, pancreatic urinary bladder, and skin premalignant, and malignant lesions.83 These mice models clearly demonstrated that in all four cancers, the expression of ARF was preceded by the activation of the DDR or p16INK4A and in fact was observed in later stages of tumor development. Moreover, several human cancer samples including, head and neck, skin, pancreas, early and stages cancers of the urinary bladder all showed similar expression profiles, as seen in mice models. A further careful analyses using epithelial and fibroblast cells have shown that the upregulation of ARF, which is delayed after the induction of DDR, requires at least two oncogenic activators. This shows a sharp contrast to situations causing a more immediate oncogenic stress causing DDR, which usually requires only one oncogene stimulation. Overall, this report, although still preliminary, has provided crucial information about the time sequence and different roles of DDR and ARF. Moreover, it has provided a possibility of two lines of defense mechanisms, where DDR can provide an initial defense over lower dose of oncogenic activation, while ARF provides a second line, later stage defense for tumor development, which is designed to defend DNA from more powerful stimuli of escalating oncogenic overload (Figure 3(a)).
A second related report by Velimezi et al. addressed the relationship, more of antagonizing partnership, between DDR and ARF pathways from a different set of experiments.84 In this report, it has been demonstrated that in human oncogene-transformed cells and cancer cell lines, ATM suppressed both protein level and activity of ARF, secondary to the kinase activity of ATM. Mechanistically, ATM promotes the degradation of ARF protein. ATM promotes the activity of protein phosphatase 1, and this in turn reduces phosphorylation of nucleophosmin (NPM). Of note, ARF is attached to NPM for its stability and once NPM is dephosphorylated by ATM, it is not able to bind NPM anymore. The detached ARF from NPM then undergoes degradation by ULF E3-ubiquitin ligase.85 This observation was further reinforced by the finding that, in resected samples from cancer patients, lack of ATM expression correlated with increased protein level of ARF. Finally, the suppression of ATM promotes the tumor-suppressive effects of p53 which is modulated by ARF. Overall, the mechanistic insight provided by this report provides an important potential explanation for timing and interplay between the DDR and ARF.
As discussed above, both reports by Evangelou et al. and Velimezi et al. widened our understanding for tumorigenesis of sporadic cancer biology and for the designing of potential biomarkers in the treatment of advanced tumors83,84 (Figure 3(a)). However, it is unclear how oncogene-induced replication stress causes various genomic instabilities, particularly for tumor suppressor/oncogenic pathway itself and certainly; it was expected that genomic instability can significantly affect the protein function related with this pathway mostly by genetic mutations. However, although various degrees of genetic change have been identified, in most of cases, functional important genetic alterations have not been described in a significant level. Instead, other mechanisms seem to play a more important role. Foremost, biochemical changes like kinase-dependent protein phosphorylation or modified protein/protein interaction and epigenetic changes like DNA methylation or histone acetylation seem to play as dynamic regulatory players for both barrier and promoters of genomic instability.85–87 Of note, so far, various genetic changes have been reported for six selected genes including TP53, MDM2, MDM4, CDKN2A, CDK4, and RB1, of which these mutation data are obtained from various sources including cell line, xenograft and resected cancer samples. While these genetic changes involve point mutations, amplification, and deletions, the details of mutational changes are all different, depending on samples from cells, cancer cell lines, and cancer tissues in various settings.
Understanding genomic instability from mutation analysis
Limitations of interpretations, mutational signatures, and unexpected discoveries
With the understanding of oncogene-induced genomic instability, it is worthwhile to look at the mutation rate of caretaker genes in some more details. The “mutator hypothesis” assumes that caretaker gene mutations are frequent and occur at the initial stage of tumorigenesis, thereby playing a crucial role for cancer development.4,12 While this hypothesis has well been explained in the case of hereditary cancers, such cancer patients are usually rare. However, in a more predominant form of cancer, sporadic cancers, all the reported DNA sequencing studies fail to identify frequently mutated genes among known or predicted DNA repair and mitotic checkpoint gene families.17–20 While we cannot rule out unlikely possibility that mutations from unknown/uncharacterized caretaker genes are not discovered yet, the infrequent rate of mutations in caretaker genes in sporadic cancers are still an enigma. Moreover, mutation frequency does not necessarily accompany frequency of inactivation of such gene products. Besides, as not all non-synonymous mutations change function of gene product, it is hard to conclude that such mutations are the driving force of human cancer development.86,87
Most, but not all,88 caretaker genes are recessive, and based on the lesson from “mutator hypothesis,” such inactivation must happen in the early stage of tumorigenesis. Then how can we explain about the role of caretaker genes during tumorigenesis? Several large-scale sequencing studies confirmed that genomic instability in many sporadic human cancers have shown that caretaker gene mutations are not common, as between 65 and 98% of cancers did not have mutations in caretaker genes in the various studies. Instead, the prevailing theory has been developed that the mutation may be only one part of gene activation, and in many cases, gene function can also be suppressed by epigenetic mechanisms.89
Most recently, a different angle of interpretation was suggested by Alexandrov et al., whose group analyzed the results from the high-throughput sequencing studies.90 In this report, rather than focusing only on whether the observed mutations inactivate caretaker genes, the authors have focused on the vast data of mutations in cancer by mutation signatures. A total of 4,938,362 mutations were obtained from 7042 cancers and then were analyzed for more than 20 unique mutational signatures. While some gene signatures are shared in many cancer types, like a signature from APOBEC family of cytidine deaminases, it also seems that depending on the different types of cancer, there are various signatures specific to a single cancer type. Moreover, there are several signatures linked to three different variables: exposures to mutagen like nicotine exposure, defects in DNA maintenance, and age of cancer patient at the time of diagnosis. In addition to various mutational signatures, small genomic regions harboring hypermutation described as “kataegis,” is universally described in many different cancer types. The results reveal not only the complexity of diverse mutational processes during human carcinogenesis but also promoted the standard of patient care from the aspects of cancer prevention and designing of optimal cancer treatment.
However, detailed mechanistic understanding about “How these signatures are developed?” are still largely lacking, while at least one report has tried address this question. Recently, unexpected oncogenic property of p21WAF1/Cip1 (p21), which is downstream effectors of p53 as a conventional tumor suppressor pathway, was described. Basically, chronic expression of p21WAF1/Cip1(p21) in a p53-deficient environment causes genomic instability. The mechanism behind this unexpected observation can be summarized as “p21 induced deregulation of the replication licensing machinery,” which happens only in the absence of p53 function.48 As a succession of this report, Galanos et al. provided more mechanistic insights for these observations by Alexandrov et al.90,91 This report basically demonstrated that p21 can suppress DNA repair pathways for nucleotide abnormalities.91 As a result, decreased 92amount of single nucleotide substitutions occur, while highly deleterious DNA DSBs increases. The result is such a way that cell populations with persistent p21 activity, when protective effects of p53 are lacking, are prone to dangerously enhanced genomic instability. This also creates a characteristic mutational signature landscape and led to the important discovery of novel DSB repair mechanism, that the DSBs are repaired by Rad52-dependent break-induced replication in addition to repair of single-strand annealing.
In summary, these observations altogether48,89,90 confirmed the importance of careful analysis of mutational signatures. It also provided an interesting possibility that mutational signatures can be used as a clue to discover the history of DNA repair, leading to genomic instability. Moreover, it demonstrated surprising observation that persistent expression of p21, a conventional tumor suppressor, can facilitate enhanced genomic instability by reshuffling the repair process. Finally, a new discovery of Rad52 as a key player of DSB repair places Rad52 to the new position of not only an important research subject of genomic instability but also potential therapeutic target.
Genetic mutations in sporadic cancer
So far, three high-throughput sequencing studies examined the coding sequences of altogether of 18,191–20,661 genes in four carcinomas. Four cancer types including colon cancer, pancreatic cancer, breast cancer, and glioblastomas25–28 are analyzed, among other genes, for mutations in various caretaker gene families. Cumulatively, in the four cancer types, 68 cancers were analyzed in the discovery phase and 221 cancers were examined in the following study either for validation or prevalence screens. There are several important observations. First, ERCC6 (RAD26), a gene involved in transcription coupled-nucleotide excision repair, was the most frequently mutated caretaker gene. However, this gene was mutated only in six cancers. Second, four genes involved in the repair of DNA DSBs, namely MRE11, PRKDC, BRCA1 and BRCA2, and one gene involved in the spindle assembly checkpoint, centromere-associated protein E, were each mutated in only two cancers. Third, other rare mutations were found in the multiple genes active in homologous recombination, DDR, and DNA repair genes. However, the mutation frequencies for all these rare mutations are very low and are not mentioned here.
Likewise, studies focused on 600 genes involved in DNA repair and cell cycle checkpoint genes are performed. From the analysis of 188 lung adenocarcinomas and 91 glioblastomas,29,30 overall mutational patterns carry similar trend to those studies done for four different cancers as discussed above. In the lung adenocarcinomas, the NHEJ DNA repair gene PRKDC and the mismatch repair gene MSH6, both of which are important caretaker genes, were mutated only in six and four cases, respectively. The three HRR genes including BAP1, BRCA1-associated RING domain 1 and BRCA2 were mutated in two cases each. Lastly, the HRR gene BRCA1, the base-exchange repair gene XRCC1, the nucleotide excision repair gene, ERCC2 were mutated in one case. Likewise, two of the mitotic checkpoint genes, BUB1 and STK12, were mutated in one case each. In summary, mutations in gatekeeper genes including DNA repair genes and checkpoint genes are rare in sporadic cancer, and therefore genomic instability in sporadic cancer cannot be explained by mutator hypothesis.
However, in the glioblastoma, there are two unexpected findings. First, among caretaker genes, mutational frequencies profoundly change between the untreated and treated cases. For example, in the 72 untreated cases, only two mutations were found: one targeting MSH2 and the other targeting BRCA, both of which are involved in DNA repair pathways. However, among treated cases, 14 mutations were found out of 26 cases. This 26-fold increase in mutation frequency for both genes are unexpected and suggests that mutations in these two caretaker genes in a glioblastoma might be a late event as a response to treatment, which often involves chemotherapy and radiation therapy. Second, and probably more importantly, the presence of mutations targeting energy controlling enzyme like isocitrate dehydrogenase 1 (IDH 1) was quite surprising. The IDH 1 encodes a metabolic enzyme responsible for the conversion of isocitrate to α-ketoglutarate and is mutated in 12% of the tested glioblastoma samples.25–28 Further biochemical analysis has demonstrated that newly identified onco-metabolite can promote growth and survival signals for glioblastomas and it needs to be seen if such onco-metabolite can play a similar role in the development of other sporadic tumors. Again, these data overall demonstrate that the low frequency of caretaker genes mutations does not support the “mutator hypothesis” for sporadic cancers. Likewise, the role of signaling pathways leading into the development of certain cancers of the lungs, head, and/or neck seems to be partially dependent on outside signals like EGF. Overall, these rather extensive mutation analyses support the theory of “few oncogenes induced genomic instabilities” as a reasonable theory for underlying mechanism of the development of sporadic cancer.
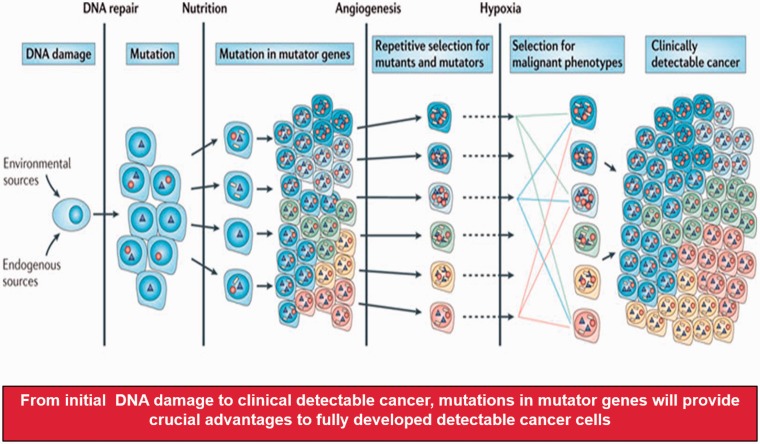
For sporadic solid tumors, epidemiological evidence suggested that as many as 20 years are required, between the time of patient’s exposure to a carcinogen like smoking and full development of cancer, enough for the radiological diagnosis. While tumor progression is suppressed by cellular defense mechanisms, several steps are required for successful cancer developments. While there are many factors that can play a role, compromised DNA repair processes, the availability of nutrition necessary for energy requirement, the requirement of accelerated angiogenesis feeding tumor cells to maximize its volume, and responses to challenging metabolic environment like hypoxia are four areas of crucial importance. In Figure 4, a series of steps are shown in a simplified manner to demonstrate this process. However, this might be one of the best ways of explaining, “How accumulated mutations can induce genomic instability and thus lead into a full-blown cancer development.” We expect that future studies will focus on the genetic and epigenetic regulatory mechanisms of gene expression, and more broadly on modifications of known oncogenic proteins such as by phosphorylation or acetylation.
Figure 4.
Accumulated mutators can lead into full blown cancer development. While various barriers to tumor progression exist, including DNA repair processes, accumulated mutators can promote the availability of securing nutrition, fulfilling the requirement of angiogenesis and thus allow the tumor to increase in size and responses to hypoxia. Circles represent mutations in genes that result in enhanced mutagenesis, triangles indicate driver mutations that are selected on the basis of changes in the tumor microenvironment and white rectangles represent passenger mutations (Modified from Loeb12).
Conclusion and future directions
In this review, we have tried to provide a comprehensive understanding for genomic instability in cancer, focused on sporadic tumors. While mutations in DNA repair genes have been well documented as a basis of genomic instability in hereditary cancers, in sporadic cancers, the molecular basis of genomic instability is less well understood. As we have extensively discussed in this review, several large-scale DNA sequencing studies confirm that, in the case of sporadic cancers, mutations in caretaker genes like DNA repair genes may not able to explain necessary steps, leading into genomic instability. An “oncogene-induced DNA replication stress” model may explain as one of the responsible mechanisms for the presence of genomic instability in sporadic cancers. While studies in MYC gene have provided mechanistic insight, other alternatives are also possible, such as interactions between key tumor cell genes and other components like metabolic environment or immune surveillance systems. Expected future works are needed for precise understanding of the molecular basis of genomic instability in sporadic cancers, while interpretation of such work products will be undoubtedly challenging. However, a comprehensive understanding of these few mutator genes or protein modifications might be crucial in the early detection of various cancers, which will be one of the only viable options for a “high chance for cure.”
Acknowledgements
CM is deeply indebted to Drs. HJ Moon and JS Yoon (both of whom unfortunately recently passed away) on their encouragement for the completion of this review. The JSY Research Institute is a research organization founded in memory of Dr Yoon, who is Dr Moon's mother and Dr Moon owns 100% of its equity.
Authors' contributions
All authors participated in the interpretation and analysis of literature reviewed; JJM and AL carried out the literature searches and writing of the manuscript. JJM and CM were involved in the writing and editing of the manuscript. CM has provided overall guidelines.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The study was supported in part by Cancer Research Grant from Pyung-Ya Foundation (to CM).
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57–70 [DOI] [PubMed] [Google Scholar]
- 2.Boveri T. Zur frage der entstehung maligner tumoren. Jena,: G. Fischer, 1914. [Google Scholar]
- 3.Von Hansemann D. Ueber asymmetrische Zelltheilung in Epithelkrebsen und deren biologische Bedeutung. Archiv f Pathol Anat 1890; 119:299–326 [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science 1976; 194:23–8 [DOI] [PubMed] [Google Scholar]
- 5.Winge Ö. Zytologische untersuchungen über die natur maligner tumoren. ZZellforsch 1930; 10:683–735 [Google Scholar]
- 6.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993; 75:1027–38 [DOI] [PubMed] [Google Scholar]
- 7.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA, Nyström L. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 1993; 75:1215–25 [DOI] [PubMed] [Google Scholar]
- 8.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat Genet 2002; 30:227–32 [DOI] [PubMed] [Google Scholar]
- 9.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 2008; 13:472–82 [DOI] [PubMed] [Google Scholar]
- 10.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science (New York, NY) 2008; 319:13525. [DOI] [PubMed] [Google Scholar]
- 11.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009; 136:823–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res 1991; 51:3075–9 [PubMed] [Google Scholar]
- 13.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nature Rev Cancer 2005; 5:564–73 [DOI] [PubMed] [Google Scholar]
- 14.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J 2003; 374:577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. Journal of Clinical Oncology: official Journal of the American Society of Clinical Oncology 2006; 24:3799–808 [DOI] [PubMed] [Google Scholar]
- 16.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet 2009; 17:722–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature 2004; 432:338–41 [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, Parsons DW, Traverso G, Awad M, Silliman N, Ptak J, Szabo S, Willson JK, Markowitz SD, Goldberg ML, Karess R, Kinzler KW, Vogelstein B, Velculescu VE, Lengauer C. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res 2004; 64:2998–3001 [DOI] [PubMed] [Google Scholar]
- 19.Cahill DP, da Costa LT, Carson-Walter EB, Kinzler KW, Vogelstein B, Lengauer C. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics 1999; 58:181–7 [DOI] [PubMed] [Google Scholar]
- 20.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature 1998; 392:300–3 [DOI] [PubMed] [Google Scholar]
- 21.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Ørntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005; 434:864–70 [DOI] [PubMed] [Google Scholar]
- 22.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Ørntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006; 444:633–7 [DOI] [PubMed] [Google Scholar]
- 23.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444:638–42 [DOI] [PubMed] [Google Scholar]
- 24.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005; 434:907–13 [DOI] [PubMed] [Google Scholar]
- 25.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science (New York, NY) 2006; 314:268–74 [DOI] [PubMed] [Google Scholar]
- 26.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science (New York, NY) 2007; 318:1108–13 [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY) 2008; 321:1801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science (New York, NY) 2008; 321:1807–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455:1069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455:1061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature 1997; 386:623–7 [DOI] [PubMed] [Google Scholar]
- 32.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol 1999; 9:M57–60 [PubMed] [Google Scholar]
- 33.Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA 1992; 89:7491–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, Vogelstein B, Lengauer C. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res 2002; 62:1129–33 [PubMed] [Google Scholar]
- 35.Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW, Xu Y. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol Cell Biol 2005; 25:661–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkovich E, Ginsberg D. ATM is a target for positive regulation by E2F-1. Oncogene 2003; 22:161–7 [DOI] [PubMed] [Google Scholar]
- 37.Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-RAS oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci USA 1994; 91:5124–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA 1999; 96:3940–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol Cell 2002; 9:1067–78 [DOI] [PubMed] [Google Scholar]
- 40.Mai S, Fluri M, Siwarski D, Huppi K. Genomic instability in MYCER-activated Rat1A-MYCER cells. Chromosome Res 1996; 4:365–71 [DOI] [PubMed] [Google Scholar]
- 41.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature 1999; 401:297–300 [DOI] [PubMed] [Google Scholar]
- 42.Tanaka S, Diffley JF. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev 2002; 16:2639–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo RA, Poon RY. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev 2004; 18:1317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet 2007; 41:169–92 [DOI] [PubMed] [Google Scholar]
- 45.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene 2008; 27:3256–64 [DOI] [PubMed] [Google Scholar]
- 46.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998; 95:29–39 [DOI] [PubMed] [Google Scholar]
- 47.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993; 366:704–7 [DOI] [PubMed] [Google Scholar]
- 48.Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ, Kokkalis A, Roumelioti FM, Gagos S, Tzetis M, Canovas B, Igea A, Ahuja AK, Zellweger R, Havaki S, Kanavakis E, Kletsas D, Roninson IB, Garbis SD, Lopes M, Nebreda A, Thanos D, Blow JJ, Townsend P, Sørensen CS, Bartek J, Gorgoulis VG1. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol 2016; 18:777–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-MYC protein. Genes Dev 2003; 17:1115–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grandori C, Cowley SM, James LP, Eisenman RN. The myc/max/mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000; 16:653–99 [DOI] [PubMed] [Google Scholar]
- 51.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 2004; 4:562–8 [DOI] [PubMed] [Google Scholar]
- 52.Collins S, Groudine M. Amplification of endogenous MYC-related DNA sequences in a human myeloid leukaemia cell line. Nature 1982; 298:679–81 [DOI] [PubMed] [Google Scholar]
- 53.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-MYC onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci 1982; 79:7824–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalla-Favera R, Gelmann EP, Martinotti S, Franchini G, Papas TS, Gallo RC, Wong-Staal F. Cloning and characterization of different human sequences related to the onc gene (v-MYC) of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci 1982; 79:6497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RSK, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001; 412:341–6 [DOI] [PubMed] [Google Scholar]
- 56.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer 2002; 2:764–76 [DOI] [PubMed] [Google Scholar]
- 57.Wade M, Wahl GM. c-MYC, genome instability, and tumorigenesis: the devil is in the details. Curr Top Microbiol Immunol 2006; 302:169–203 [DOI] [PubMed] [Google Scholar]
- 58.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell 2005; 123:13–24 [DOI] [PubMed] [Google Scholar]
- 59.Vennstrom B, Sheiness D, Zabielski J, Bishop JM. Isolation and characterization of c-MYC, a cellular homolog of the oncogene (v-MYC) of avian myelocytomatosis virus strain 29. J Virol 1982; 42(3):773–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev 2000; 14:1553–77 [PubMed] [Google Scholar]
- 61.Kolli S, Buchmann AM, Williams J, Weitzman S, Thimmapaya B. Antisense-mediated depletion of p300 in human cells leads to premature G1 exit and up-regulation of c-MYC. Proc Natl Acad Sci 2001; 98:4646–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajabi HN, Baluchamy S, Kolli S, Nag A, Srinivas R, Raychaudhuri P, Thimmapaya B. Effects of depletion of CREB-binding protein on c-MYC regulation and cell cycle G1-S transition. J Biol Chem 2005; 280:361–74 [DOI] [PubMed] [Google Scholar]
- 63.Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 2010; 11:728–38 [DOI] [PubMed] [Google Scholar]
- 64.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 2009; 21:771–7 [DOI] [PubMed] [Google Scholar]
- 65.Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci 2011; 36:405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA 2009; 106:20240–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 2011; 18:471–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011; 146:931–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 2006; 8:358–66 [DOI] [PubMed] [Google Scholar]
- 70.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 2010; 37:247–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 2006; 21:581–7 [DOI] [PubMed] [Google Scholar]
- 72.Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011; 146:80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 2010; 24:1208–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase CDK. Embo J 1998; 17:5699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. BioEssays 2003; 25:116–25 [DOI] [PubMed] [Google Scholar]
- 76.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 2002; 277:33049–57 [DOI] [PubMed] [Google Scholar]
- 77.Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 2011; 21:2055–63 [DOI] [PubMed] [Google Scholar]
- 78.Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 2012; 19:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-MYC. Nature 2007; 448:445–51 [DOI] [PubMed] [Google Scholar]
- 80.Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PloS One 2011; 6:e17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of MYC-dependent DNA replication stress. Cell Rep 2013; 3:1629–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev 2003; 194:77–95 [DOI] [PubMed] [Google Scholar]
- 83.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res 2003; 63:1798–805 [PubMed] [Google Scholar]
- 84.Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, Kosar M, Liloglou T, Trougakos IP, Dyrskjot L, Andersen CL, Papaioannou M, Drosos Y, Papafotiou G, Hodny Z, Sosa-Pineda B, Wu XR, Klinakis A, Ørntoft T, Lukas J, Bartek J, Gorgoulis VG. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell Death Differ 2013; 20:1485–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K, Kotsinas A, Orsolic I, Bursac S, Cokaric-Brdovcak M, Zoumpourlis V, Kletsas D, Papafotiou G, Klinakis A, Volarevic S, Gu W, Bartek J, Halazonetis TD, Gorgoulis VG. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol 2013; 15:967–77 [DOI] [PubMed] [Google Scholar]
- 86.Enomoto T, Lindstrom MS, Jin A, Ke H, Zhang Y. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability. J Biol Chem 2006; 281:18463–72 [DOI] [PubMed] [Google Scholar]
- 87.Lee W, Yue P, Zhang Z. Analytical methods for inferring functional effects of single base pair substitutions in human cancers. Hum Genet 2009; 126:481–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parmigiani G, Boca S, Lin J, Kinzler KW, Velculescu V, Vogelstein B. Design and analysis issues in genome-wide somatic mutation studies of cancer. Genomics 2009; 93:17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Driscoll M. Haploinsufficiency of DNA damage response genes and their potential influence in human genomic disorders. Curr Genom 2008; 9:137–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148–59 [DOI] [PubMed] [Google Scholar]
- 91.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC Mmml-Seq Consortium ICGC PedBrain Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature 2013; 7463:415–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galanos P, Pappas G, Polyzos A, Kotsinas A, Svolaki I, Giakoumakis NN, Glytsou C, Pateras IS1, Swain U, Souliotis VL, Georgakilas AG, Geacintov N, Scorrano L, Lukas C, Lukas J, Livneh Z, Lygerou Z, Chowdhury D, Sørensen CS, Bartek J, Gorgoulis VG. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol 2018; 19:1401–9 [DOI] [PMC free article] [PubMed] [Google Scholar]