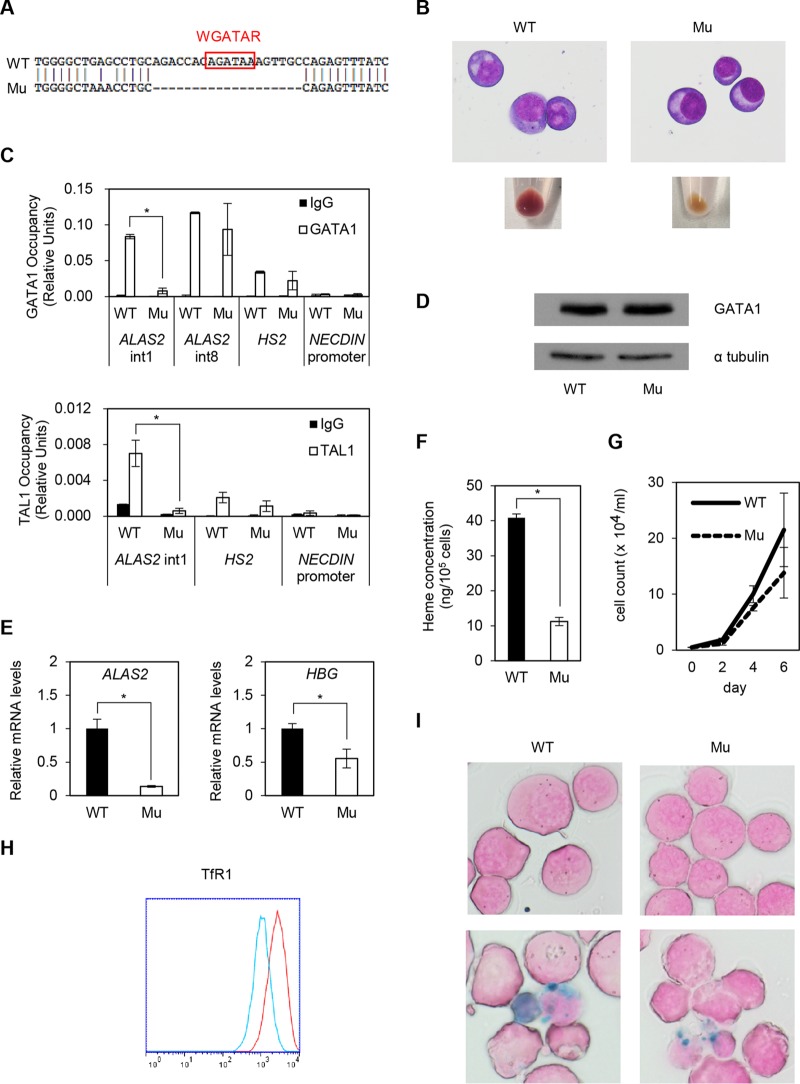
FIG 4.
Establishment of XLSA clones by CRISPR/Cas9-mediated ablation of ALAS2 cis elements in HiDEP cells. (A) Sequence analysis. WT, wild-type HiDEP cells; Mu, XLSA clones. (B) May-Giemsa staining (upper) and cell pellets (lower). (C) Quantitative ChIP analysis to detect endogenous GATA-1 (upper) and TAL1 (lower) chromatin occupancy in wild-type HiDEP cells and in the XLSA clone cells. HS2, which is the DNase I hypersensitivity site at the β-globin locus control region (6), and the NECDIN promoter were used as positive and negative controls, respectively. Data are expressed as means ± SD (n = 3). (D) Western blot to detect GATA-1 in wild-type HiDEP cells and XLSA clone cells. α-Tubulin was used as a loading control. Representative data from at least two independent experiments are shown. (E) Quantitative RT-PCR analysis for ALAS2 and HBG expression in wild-type HiDEP cells and XLSA clone cells. Values presented are relative to those of GAPDH mRNA. Data represent averages from three independent experiments and are expressed as means ± SD. *, P < 0.05. (F) Intracellular heme concentration in wild-type HiDEP cells and XLSA clone cells. Data are presented as means ± SD (n = 3). *, P < 0.05. (G) Changes in total cell number by CRISPR/Cas9-mediated ablation of ALAS2 cis elements at intron 1 in HiDEP cells. (H) FACS analysis to evaluate TfR1 expression in wild-type HiDEP cells and XLSA clone cells. Representative data from at least two independent experiments are shown. Red, wild type; blue, XLSA clone cells. (I) Prussian blue staining (upper, without SFC; lower, with 100 μM SFC for 4 days) of wild-type HiDEP cells and XLSA clone cells.