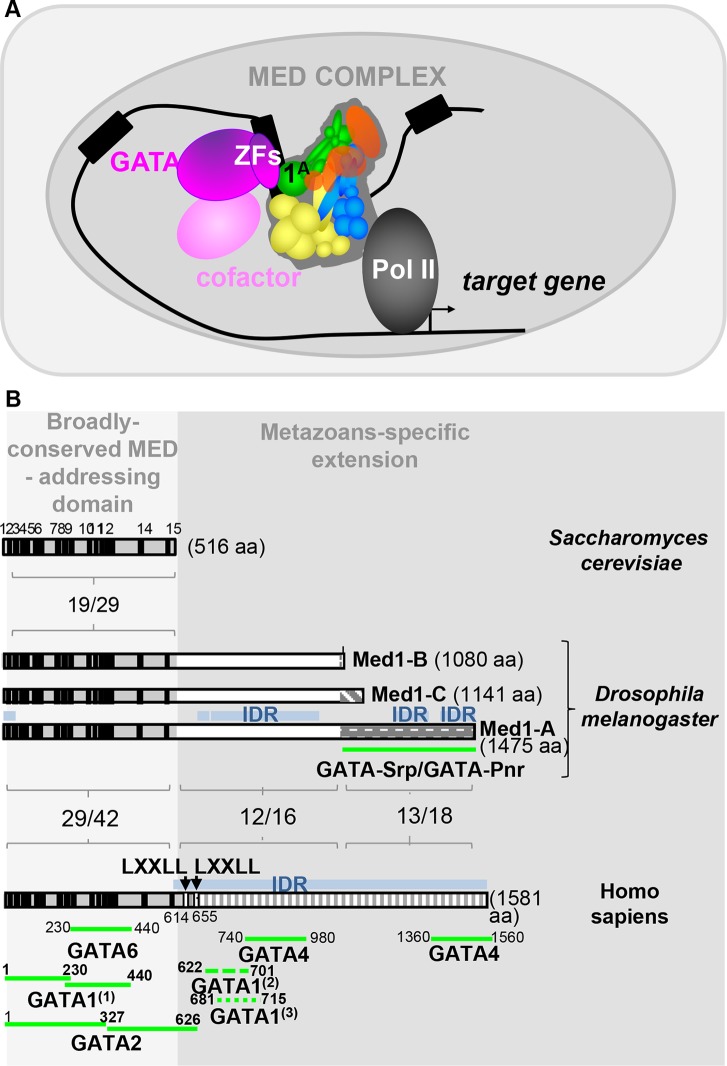
FIG 7.
MED1-GATA molecular partnership. (A) Model of Med1-GATA partnership. GATA transcription factors are represented by fuchsia, with GATA binding domains indicated as ZF, and corresponding cofactors are in light pink. Mediator complex is composed of four modules, tail (yellow), head (blue), CDK8 (red), and middle (green), with the latter including the Med1 subunit isoforms. (B) Med1-GATA interacting domains in Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens. The light gray part emphasizes their N-terminal region, comprising 15 short evolutionarily conserved motifs (black boxes), which have been shown to be sufficient for Med1 integration into the mammalian MED complex. Conversely, the darker gray part emphasizes the divergent long metazoan-specific extensions. Percentage of identity/similarity between homologous protein domains is indicated. Blue boxes represent intrinsically disordered regions (IDR) for which no conserved motifs could be identified. Their richness in serine residues (21% to 22%) accounts for most of the amino acid identity observed in the metazoan-specific extension. Position of GATA-interacting domains identified to date for human GATA6 (6), GATA2 (9), GATA4 (7), and GATA1 (55) and Drosophila Srp and Pnr (this work) are indicated below. In the case of mammalian GATA1, note that two independent analyses from Stumpf et al. (7) and Mizuta et al. (55) identified two overlapping domains at the beginning of the divergent metazoan-specific extension, whereas Crawford and collaborators (6) identified two other regions within the conserved and structured N-terminal part that revealed some discrepancies.