Figure 3. Kinase motifs.
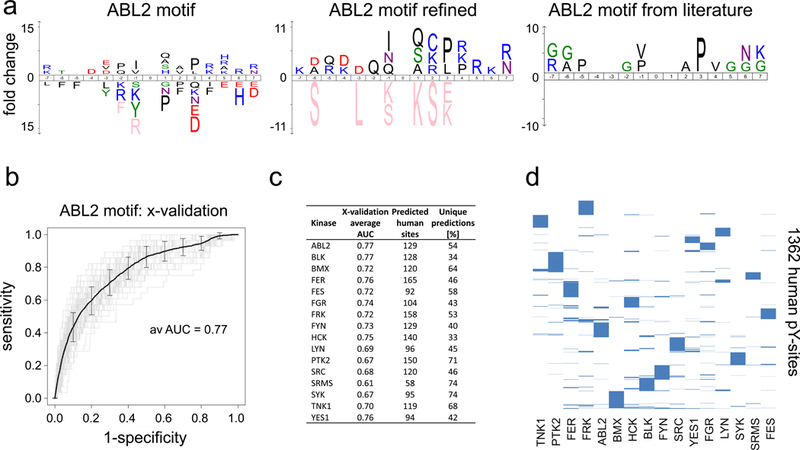
a. Motif generation. Using ico-Logo (Colaert et al., 2009) linear sequence motifs covering 7 amino acids N- and and C-terminal of the phosphorylated tyrosine residues (position 0, Y not shown). Novel motifs were generated for all 16 NTRKS and compared to the literature in ROC analyses (Suppl. Figure 2), ABL2 motifs are exemplarily shown. b. 10-fold cross-validation for kinase motifs derived from yeast pY-sites resulted AUC values in the range of 0.66–0.78. The ROC curve for ABL2 motif is exemplarily shown (see Suppl. Figure 3a). c. Summary of motif analyses and motif predictions for human phospho-tyrosine sites. AUC values from the cross-validation (x-validation average AUC) are listed. At an accuracy cut-off of 0.995 between 165 and 92 human pY-sites were scored (Predicted human sites). About 50% of the motif based kinase-substrate assignments are unique for a single kinase (Unique predictions). d. Human pY-sites with motif based kinase assignments. Graphical representation showing motif based kinase-substrate relationships for 1362 human phospho-tyrosine sites.
