Figure 8. Structural representation of predicted binding modes of the identified GLUT inhibitors.
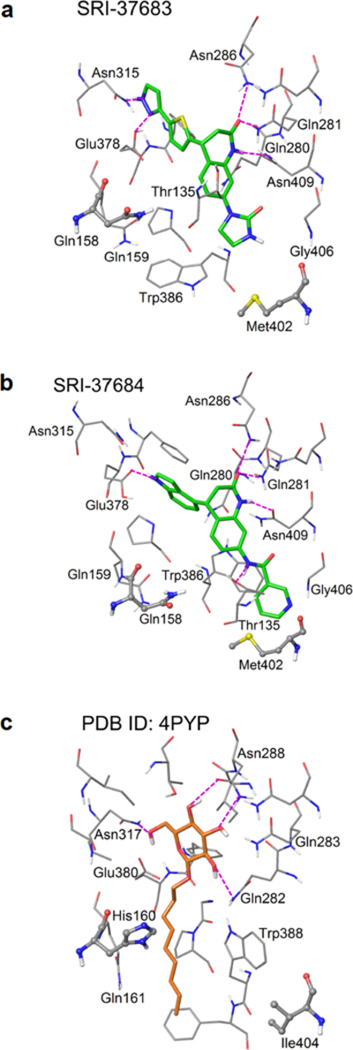
The predicted binding modes of (a) SRI-37683 and (b) SRI-37684 in the inward-open conformation of the modeled GLUT3 crystal structure. (c) The ligand (n-nonyl-β-D-glucopyranoside) binding mode in the crystal structure of GLUT1 in the inward-open conformation (PDB ID: 4PYP). Ligand molecules are represented in slide sticks. Binding site residues are shown in thin tubes. The two residues that are not conserved between GLUT1 and GLUT3 are illustrated in stick & ball mode. Hydrogen bonds are shown in dashed purple lines.
