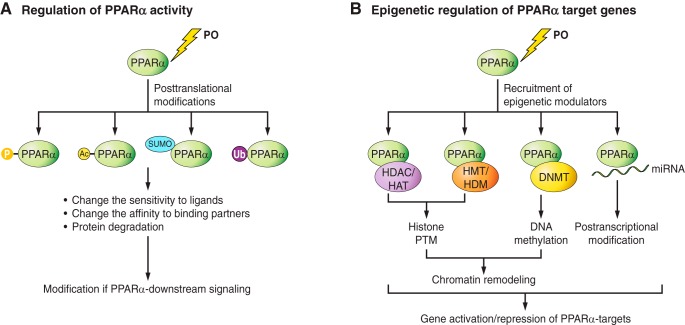
Fig. 2.
Hypothetical regulatory mechanisms of PPARα activity through posttranslational modifications (PTMs, A) and epigenetic regulation (B). A: during PO, PPARα undergoes various PTMs, including phosphorylation (P), Ac, SUMOylation [small ubiquitin-like modifier (SUMO)ylation], and ubiquitination (Ubi). Consequently, PPARα changes in its sensitivity to ligands and/or its affinity for its binding proteins, leading to the modifications in PPARα downstream signaling. Ubiquitination of PPARs can result in protein degradation. B: PO induces the recruitment of epigenetic modulators to the promoter regions of PPARα target genes. Depending on which modulators are recruited, PPARα can trigger histone PTMs, DNA methylation, or posttranscriptional modifications. In general, histone acetylation leads to gene activation, whereas histone deacetylation leads to gene repression. The functional consequence of histone methylation is complex, since transcriptional control depends on the position of the lysine residue that undergoes methylation. Direct DNA methylation by methyltransferase (DNMTs) generally leads to gene silencing. A micro-RNA (miR) is a small noncoding RNA molecule that may alter expression of PPARα targets. HAT, histone acetylase; HDAC, histone deacetylase; HMT, histone methyltransferase; HDM, histone demethylase.