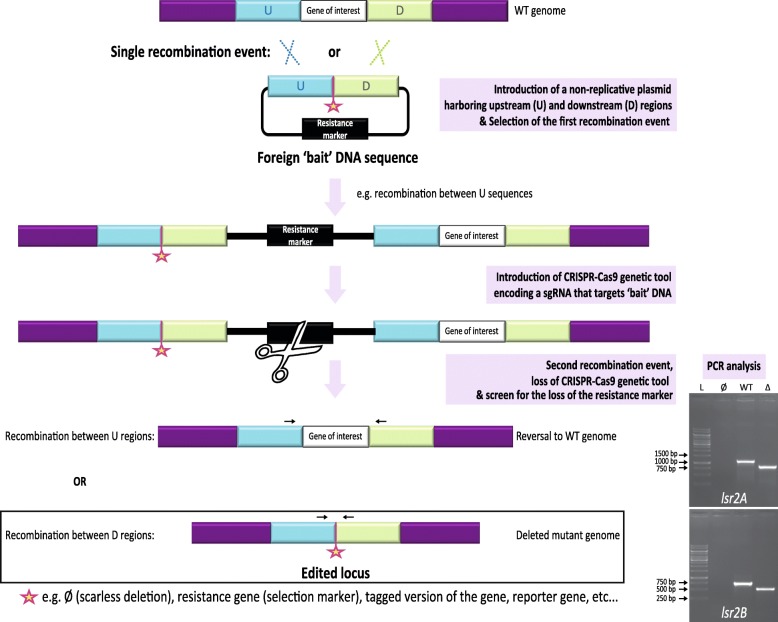
Fig. 1.
Main stages of the CRISPR-Cas9 targeting ‘bait’ DNA approach to perform gene-copy specific editing. The approach is based on the specific insertion by homologous recombination of a selection marker and a foreign DNA harboring the upstream (‘U’) and downstream (‘D’) regions surrounding the copy of interest. The star represents the various possible designs of the model DNA used to repair the lesion by homologous recombination. For clarity reasons, the scheme only represents a first recombination event occurring between U sequences but the homologous recombination could also take place between D sequences. Thereafter a genetic tool encoding Cas9 and a sgRNA specifically directed against foreign ‘bait’ DNA is introduced transitorily in the recombinant strain. The double strand break induced by this complex can be repaired by a second recombination event between either the U or D sequences, resulting in reversion or in the copy-specific editing, respectively. In absence of a second selection marker at the star position, the distinction between both situations can be achieved by PCR (arrows represent primer positions). The pictures show representative results of PCR performed without template (“Ø”), on the native genome (“WT”) or on the genome of scarless deleted mutants (“Δ”) obtained during this study. The PCR product expected sizes were 1106 pb and 815 pb for native and deleted lsr2A loci, and 729 pb and 481 pb for native and deleted lsr2B loci, respectively (see Method section for further details). The PCR product expected sizes for native and deleted loci are indicated (“L”: GeneRuler™1 kb DNA ladder, Thermo Scientific)