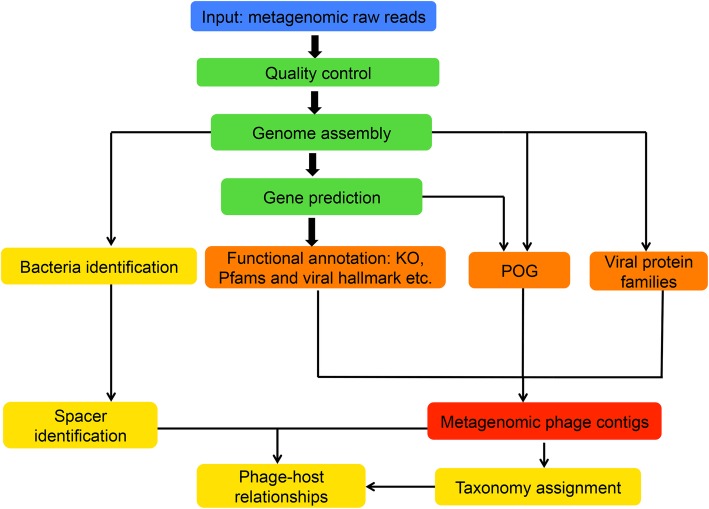
Fig. 1.
The workflow of VirMiner. The high-quality reads are assembled into contigs using IDBA_UD [33]. HMM model from GeneMark [34] was used for gene prediction. Functional profiles are generated by searching against different databases including KO, Pfam, and viral protein families defined by Paez-Espino et al. [24], POG 2012, and uPOGs. The R package randomForest was employed to identify phage contigs. The taxonomy affiliations of identified phage contigs are identified using the RDP classifier. Phage-host interaction prediction was performed using the CRISPR-spacer based method