Abstract
Background
Cross protection is a promising alternative to control plant viral diseases. One critical factor limiting the application of cross protection is the availability of attenuated mutants or mild strains. Potato virus X (PVX) infects many crops and induces huge economic losses to agricultural production. However, researches on the variability and mechanism of PVX virulence are scarce.
Methods
The mutants were obtained by introducing mutations into the RNA dependent RNA polymerase (RdRp) gene of PVX via site-directed mutagenesis. Attenuated mutants were screen according to their symptoms in Nicotiana benthamiana plants. The protection efficacy against severe infection were evaluated with interval of 5, 10 and 15 days.
Results
Among the 40 mutants obtained, four mutants carrying substitutions of either Glu46, Asn863, Asn968 or Glu1001 to Ala in PVX RdRp showed drastically attenuated symptom, accompanying with reduced accumulation levels of coat protein, plus- and minus-sense RNAs. When the interval between protective and challenging inoculations was 15 days, mutant E1001A (with substitution of Glu1001 to Ala in RdRp) provided complete protection against severe infection in both Nicotiana benthamiana and tomato, while E46A (Glu46 mutated to Ala) provided incomplete protection. To reduce the risk of reverse mutation, we constructed mutant dM which carries double mutations of both Glu46 and Glu1001 to Ala in RdRp. The mutant dM could provide effective protection against severe PVX infection.
Conclusion
Mutations of Glu46, Asn863, Asn968 or Glu1001 to Ala in PVX RdRp significantly reduced the viral symptoms. Mutants E1001A and E46A could provide effective protection against wild type PVX in both Nicotiana benthamiana and tomato. These results provide theoretical and practical bases for the control of PVX via cross protection.
Keywords: Cross protection, Potato virus X, RNA dependent RNA polymerase, Virulence
Background
Cross protection is a phenomenon in which plants infected or pre-inoculated with one virus with mild symptoms will show resistance to the subsequent infection by a severe isolate of the same or closely related virus [1–3]. It was first described with Tobacco mosaic virus (TMV) by McKinney in 1929 [4]. Since then, cross protection has been used to control viruses including Bean yellow mosaic virus [5], Citrus tristeza virus [6, 7], Clover yellow vein virus [5], Cucumber green mottle mosaic virus [8], Cucumber mosaic virus [9–11], East African cassava mosaic virus-Uganda [12], Papaya ringspot virus [13, 14], Pepipo mosaic virus [15], Pepper mild mottle virus [16], Soybean mosaic virus [17], Tobacco mosaic virus [5, 18], Tomato mosaic virus [2, 19–22], Watermelon mosaic virus [9], and Zucchini yellow mosaic virus [9, 23, 24].
The pre-inoculated strains used for protective inoculation have been selected from naturally occurring isolates that cause mild symptoms [6, 9], by random or direct mutagenesis of wild severe strains using nitrous acid or ultraviolet irradiation [8, 21, 22], and by cultivation at higher or lower temperature than the optimal [5, 19, 23]. However, it is time consuming to screen the mild strains for cross protection. With the advent of reverse genetics, one can obtain mutants with site-directed mutagenesis, and then study their phenotypes and evaluate their potential in cross protection [16, 24]. This provides a faster, more effective and controllable way for screening attenuated strains.
Potato virus X (PVX; genus Potexvirus, family Alphaflexiviridae) can infect a wide range of major crops in the family Solanaceae including tomato, potato, pepper and tobacco. It can be transmitted by mechanical inoculation and contact between plants [25]. PVX has a positive-sense single-stranded (ss) RNA of 6435 nucleotides (nt), with a cap at the 5′-end and poly(A) tail at the 3′-end, respectively [26–29]. Its genome contains five open reading frames (ORFs). ORF1 encodes the replication associated protein RNA-dependent RNA polymerase (RdRp) [26, 30], while the overlapping ORFs 2, 3 and 4 encode the triple gene block (TGB) proteins (TGBp1, TGBp2 and TGBp3) which are essential for virus movement [31, 32]. ORF5 encodes the coat protein (CP) [33]. PVX isolates are classified into four groups based on their responses in the potato cultivars carrying Nb, Nx or Rx genes [34, 35], and two molecular groups, Eurasia and America, based on their complete genomic sequences [28]. Planting resistant cultivars is the most economic and effective way to control PVX. However, breeding virus resistant cultivars is time consuming and the resistance could be overcome by single amino acid mutation in CP or emerging of new strains [36, 37]. Therefore, cross protection become a promising alternative strategy for PVX control.
One prerequisite for cross protection is the availability of mild strains. Salaman (1933) reported the application of mild strains in cross protection against PVX [38]. However, no PVX mild strain is available in practice. Moreover, the mechanisms regulating the virulence of PVX are largely unknown. There are several reports in which viral symptom determinants have been mapped to the silencing suppressors [24, 39–43] or the replicases [44–47]. Mutation in these proteins may abolish the RNA silencing suppressor activity and reduce viral symptoms. Some mutants carrying such mutations were reported to confer cross protection against parental viral strain [24, 48]. The 25 kDa TGBp1 (P25) is an RNA silencing suppressor and plays an important role in the movement of PVX [31, 32, 49]. We firstly tried to screen attenuated PVX mutants by introducing mutation to TGB region encoding P25. However, all the mutants in TGBp1 either had movement defective or displayed severe symptom as severe as wild type PVX. Therefore, we failed to obtain any mild strain from TGBp1 mutants. In this paper, we obtained four attenuated PVX mutants by introducing single amino acid mutation to RdRp via site-directed mutagenesis, evaluated their potential in cross protection and elucidated the underlying mechanism; we also obtained one mutant with double mutations at two amino acid sites to increase its safety.
Methods
Mutant construction
The amino acid residues (aa) predicted by software I-TASSER [50] to be exposed on the surface of PVX RdRp were substituted with aa of opposite polarity. The asparagine residue in postulated glycosylation site was mutated to alanine. Mutations were introduced to wild type PVX infectious clone pCaPVX100 by site-directed mutagenesis using Phusion high-fidelity DNA polymerase (Thermo, Finland) and primers designed following the strategy reported previously (Table 1) [51, 52]. The fidelity of all mutants was verified by sequencing.
Table 1.
Primers used for site-directed mutagenesis of PVX mutants
| Mutant | Primer | Sequences |
|---|---|---|
| V45R | Forward | 5′-CTCAAACGCGTGAAGCGGCTAATGATCTAGAGGGGTTCGGCATAG-3′ |
| Reverse | 5′-GCCGCTTCACGCGTTTGAGCGTACGGGTTAGATAGTTTGTGTTTTTCC-3′ | |
| E46A | Forward | 5′-TCAAACGGTTGCCGCGGCTAATGATCTAGAGGGGTTCGGCATAGC-3′ |
| Reverse | 5′-ATTAGCCGCGGCAACCGTTTGAGCGTACGGGTTAGATAGTTTGTGTTTTTC-3′ | |
| T140A | Forward | 5′-ACAGAGATCGCCACGGACACAGCATACATTAGTGACACTCTGCACTTC-3′ |
| Reverse | 5′-GTGTCCGTGGCGATCTCTGTGAGCTTGTCTATTATTGTTTCCTTTGGG-3′ | |
| D200A | Forward | 5′-TACTTTGGAGCCGGTTTCCAGTATATACCAGGCAACCATGGTGG-3′ |
| Reverse | 5′-TACTGGAAACCGGCTCCAAAGTATTTGAGGCTGTATATGTTCGGGT-3′ | |
| N243A | Forward | 5′-CTCGGACATCTCGCCTACACGACTGAGCAGGTTGAGATGCACACAG-3′ |
| Reverse | 5′-AGTCGTGTAGGCGAGATGTCCGAGAAAGCTATCCTTGGGGTCCCTC-3′ | |
| K299A | Forward | 5′-ATCTTTCTCCCGGCAGTTCACAACTGCAAGAAGCCGATTCTGAAGA-3′ |
| Reverse | 5′-GTTGTGAACTGCCGGGAGAAAGATCTGTGGTGGAATCACATACCTGTCA-3′ | |
| V300R | Forward | 5′-CTCCCGAAACGCCACAACTGCAAGAAGCCGATTCTGAAGAAAAC-3′ |
| Reverse | 5′-CAGTTGTGGCGTTTCGGGAGAAAGATCTGTGGTGGAATCACATACC-3′ | |
| S341R | Forward | 5′-ATTAAATCGCGCGACTTGGACAAATACTCTGCTGTGGAACTGGTTTAC-3′ |
| Reverse | 5′-GTCCAAGTCGCGCGATTTAATTAATTGTCTGACTTTGGCAAAAATG-3′ | |
| D344A | Forward | 5′-GTCTGACTTGGCGAAATACTCTGCTGTGGAACTGGTTTACTTAGTGAGC-3′ |
| Reverse | 5′-GCAGAGTATTTCGCCAAGTCAGACGATTTAATTAATTGTCTGACTTTGGCA-3′ | |
| E446A | Forward | 5′-TCGGACGTAGCCGAAATGGAAAGTTTGTTCTCAGATGGGGACCTG-3′ |
| Reverse | 5′-TTTCCATTTCGGCTACGTCCGACACTTCCCTTGGTCGGAAGG-3′ | |
| A494K | Forward | 5′-ATTAAAGAACCTAAAGGAGACAGAAATCAATACTCAAACCCTGCGGAA-3′ |
| Reverse | 5′-TTCTGTCTCCTTTAGGTTCTTTAATTCCTTGACCGACATCCATCTCA-3′ | |
| K525A | Forward | 5′-CAGACCACAGCGAAGGCTAAGCGCCTAGCTGAAATCCAGGAGTCC-3′ |
| Reverse | 5′-GCTTAGCCTTCGCTGTGGTCTGATGTTTCACCTCTCTACTGTGTTTCCTGT-3′ | |
| K526A | Forward | 5′-CAGACCACAAAGGCGGCTAAGCGCCTAGCTGAAATCCAGGAGTCC-3′ |
| Reverse | 5′-GCTTAGCCGCCTTTGTGGTCTGATGTTTCACCTCTCTACTGTGTTTCCTGT-3′ | |
| K528A | Forward | 5′-AAAGAAGGCTGCGCGCCTAGCTGAAATCCAGGAGTCCATGA-3′ |
| Reverse | 5′-TAGGCGCGCAGCCTTCTTTGTGGTCTGATGTTTCACCTCTCTACTG-3′ | |
| E540A | Forward | 5′-ATGAGAGCAGCCGGTGAAGCTGAATCAAATGAGATGAGCGGG-3′ |
| Reverse | 5′-AGCTTCACCGGCTGCTCTCATGGACTCCTGGATTTCAGCTAGGC-3′ | |
| S545A | Forward | 5′-GAAGCAAATGAGATGAGCGGGGGCATGGGGGCAATACCG-3′ |
| Reverse | 5′-CCCGCTCATCTCATTTGCTTCAGCTTCACCTTCTGCTCTCATGGACT-3′ | |
| S565A | Forward | 5′-AGCACGGCTGATGCTAGACAAGAACTCACACTCCCAACCACC-3′ |
| Reverse | 5′-GTCTAGCATCAGCCGTGCTGGGAAGTTCAGCGTTGCTCG-3′ | |
| D566A | Forward | 5′-AGCACGAGTGCCGCTAGACAAGAACTCACACTCCCAACCACCAA-3′ |
| Reverse | 5′-TCTTGTCTAGCGGCACTCGTGCTGGGAAGTTCAGCGTTGCTCG-3′ | |
| M610K | Forward | 5′-GAGACAGCAAAACAACAAGTCATCGAAGGACTCCCTTGGAAA-3′ |
| Reverse | 5′-ACTTGTTGTTTTGCTGTCTCAACAGCTTCTTTTCCAGGGAGCTT-3′ | |
| K620A | Forward | 5′-ACTCCCTTGGGCACACTGGATTCCTCAACTAAACGCTGTTGGATTC-3′ |
| Reverse | 5′-AATCCAGTGTGCCCAAGGGAGTCCTTCGATGACTTGTTGCATTG-3′ | |
| N641A | Forward | 5′-GGGATAGGGCTGGAACAATGATCATGCCTATCACAGAAATGG-3′ |
| Reverse | 5′-ATTGTTCCAGCCCTATCCCTCTGAATTTCTAGCGCCTTGAATC-3′ | |
| E662A | Forward | 5′-ACTTCCCGGCCGGAACTCCAAAAGAGTTGGCACGAGAATTGCTC-3′ |
| Reverse | 5′-TGGAGTTCCGGCCGGGAAGTCCTCTTTTTCCAACCCGGAGA-3′ | |
| N676A | Forward | 5′-TGCTCGCTATGGCGAGAAGTCCTGCCACCATCCCTTTGGAC-3′ |
| Reverse | 5′-GGACTTCTCGCCATAGCGAGCAATTCTCGTGCCAACTCTTTTGGA-3′ | |
| K773A | Forward | 5′-AGATTGGAGTGCGAAAGTGCCCAACACTGAACCATACATGTTCAAGA-3′ |
| Reverse | 5′-GGCACTTTCGCACTCCAATCTAGCCGCAGTTCATTGGTCG-3′ | |
| N777A | Forward | 5′-AAAGTGCCCGCGACTGAACCATACATGTTCAAGACCTATGAAAAGGCAT-3′ |
| Reverse | 5′-GGTTCAGTCGCGGGCACTTTCTTACTCCAATCTAGCCGCAGTTC-3′ | |
| E779A | Forward | 5′-CCAACACTGCCCCATACATGTTCAAGACCTATGAAAAGGCATTAATTGG-3′ |
| Reverse | 5′-AACATGTATGGGGCAGTGTTGGGCACTTTCTTACTCCAATCTAGCCG-3′ | |
| N863A | Forward | 5′-CGATACTATCTCGCCGCCACACACCGCAACAAGAAAGACCTTGC-3′ |
| Reverse | 5′-TGTGTGGCGGCGAGATAGTATCGGCAGTATTTTGAGAAGTACTCTGTCGC-3′ | |
| N917A | Forward | 5′-ACCGGAAGGGCCGACACGTTCACATACGCTGGATGCCAAG-3′ |
| Reverse | 5′-ACGTGTCGGCCCTTCCGGTGCCCATGTACAGCTTTCTCTTTTCA-3′ | |
| N964A | Forward | 5′-CACTTCGTGGCGACAAGTGCAAACTCTTCGGCCTTCTGGGAA-3′ |
| Reverse | 5′-TGCACTTGTCGCCACGAAGTGAATCCTGTCGGTAGCTCTAGAGAGTGC-3′ | |
| N968A | Forward | 5′-CACAAGTGCAGCGTCTTCGGCCTTCTGGGAAAAGTTAGACA GCACCC-3′ |
| Reverse | 5′-GCCGAAGACGCTGCACTTGTGTTCACGAAGTGAATCCTGTCGGT-3′ | |
| E1001A | Forward | 5′-AGCCGGCAGCCGTAGAGCCAATTCGAGAGCCTGAGCCC-3′ |
| Reverse | 5′-GCTCTACGGCTGCCGGCTCGTACTCCTTGAGTGCTTGTTCTCTC-3′ | |
| R1006A | Forward | 5′-AGCCAATTGCCGAGCCTGAGCCCCAAACACACATGTGTGT-3′ |
| Reverse | 5′-TCAGGCTCGGCAATTGGCTCTACCTCTGCCGGCTCGTACTC-3′ | |
| E1041A | Forward | 5′-ATCCACTCTGCCTCCCATGGCCATTCAAACTGTGTCCAAACTGA-3′ |
| Reverse | 5′-CCATGGGAGGCAGAGTGGATCTCTCTGTCAAACTTTTCCAAAAGTTCC-3′ | |
| S1042A | Forward | 5′-ACTCTGAAGCGCATGGCCATTCAAACTGTGTCCAAACTGAAGACACA-3′ |
| Reverse | 5′-AATGGCCATGCGCTTCAGAGTGGATCTCTCTGTCAAACTTTTCCAAAAG-3′ | |
| K1065A | Forward | 5′-TCAACAAGCAGCGGATGAGACCCTCCTCTGGGCGACCATAGATG-3′ |
| Reverse | 5′-GGTCTCATCCGCTGCTTGTTGATGCGAAAA CAGCTGGATGGTT-3′ | |
| S1092A | Forward | 5′-AATTCTTGGCCAAGAAGGACATTGGAGACGTCCTGTTTCTAAAC-3′ |
| Reverse | 5′-GTCCTTCTTGGCCAAGAATTCTCGGAAGTTTGTTTCTTGATTG-3′ | |
| N1143A | Forward | 5′-AACTTGATTGCGGGGACTGTGAGACAGAGCCCAGACTTTGAT-3′ |
| Reverse | 5′-CACAGTCCCCGCAATCAAGTTGCACTTTGACTTACTGAGGTATTTG-3′ | |
| D1348A | Forward | 5′-ACAGGCTTGAAGCGAAATTACTCCTCAAGTCGAAGCCTGTAATCACG-3′ |
| Reverse | 5′-GGAGTAATTTCGCTTCAAGCCTGTGGAAACTAAGCTTCACTTCTGGAACG-3′ | |
| K1427A | Forward | 5′-TTGTTCGATGAGGCGCAGTGTCAGGCACATACACTCACTTGCAG-3′ |
| Reverse | 5′-ACACTGCGCCTCATCGAACAAGTCATGCAGAGAGTCCTTGTGGT-3′ | |
| L1456K | Forward | 5′-CAGAAACTTTAAGTAACCGTTAAGTTACCTTAGAGATTTGAATAAGATGG-3′ |
| Reverse | 5′-TAACGGTTACTTAAAGTTTCTGAGGCGGGGAAGTGAGACAGTGCCT-3′ |
Mutated nucleotides are shown in bold type
Plant growth and virus inoculation
Plants of N. benthamiana, N. tabacum and tomato (Solamum lycopersicum) were maintained in a growth chamber with a 16 h light/8 h dark cycle and relative humidity of 70% at 23 ± 2 °C.
The infectious clone pCaPVX100 and its mutants were inoculated via the method previously described [52]. The Agrobacterium tumefaciens GV3101 cells were transformed following the freeze and thaw transformation procedure and cultured on solid LB medium at 28 °C. Agroinfiltration was performed with agrobacterium culture that was diluted in induction solution (10 mmol/L MES, pH 5.8, 0.15 mmol/L acetosyringone and 10 mmol/L MgCl2) to final optimal density of OD600 = 0.5. The first and second fully expanded leaves of 3- to 5-leaf stage N. benthamiana, N. tabacum and tomato plants were agro-infiltrated using a 1-mL needle-less syringe. From the sixth day post agro-infiltration (dpai), symptoms on the systemic leaves were recorded daily. Each mutant was inoculated to six plants and the experiments were repeated three times independently.
Western blotting analysis
The accumulation of wild type and mutant PVX in N. benthamiana and tomato plants were determined using Western blotting. Total proteins were extracted from the systemic leaves of N. benthamiana and tomato plants, separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to a nitrocellulose (NC) membrane. The NC membrane was blocked with 5% defatted milk powder in pH 7.6 Tris-buffered saline containing 0.05% Tween-20 (TBST) for 1 h, incubated in antiserum against PVX CP diluted at 1:1000 (V/V) for 1 h, followed by 1 h incubation with alkaline phosphatase conjugated goat anti-rabbit IgG diluted in 1:50000 (Sigma) and finally colorized with NBT and BCIP.
Qualitative detection of PVX
The presence of wild type and mutant PVX in plants was determined by reverse transcription-polymerase chain reaction (RT-PCR). Total RNAs were extracted from systemic leaves at 10 dpai using Transzol reagent (Transgen) according to the manufacturer’s instruction. The first-strand cDNA was synthesized using oligo (dT) primer and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Transgen). The cDNA products were used to amplify the PVX cp gene with specific primers PVXCP-F (5′-ATT GAG GCT ATC TGG AAG GA-3′) and PVXCP-R (5′-GCC TCA GCG GTT GTT GTT CC-3′). The PCR program included an initial denaturation at 94 °C for 5 min, 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C, followed by a final extension of 10 min at 72 °C. The resultant PCR products were separated by electrophoresis on 1.0% agarose gel.
Cross protection assay
To examine whether the attenuated mutants could provide protection against the wild type PVX, plants of N. benthamiana were agro-infiltrated with suspension of Agrobacterium cells containing the attenuated mutants. The plants infiltrated with Agrobacterium cells carrying empty vector pCAMBIA0390 were used as control. At 5, 10 and 15 days after the protective inoculation, the first fully expanded systemic leaves of the protected N. benthamiana plants were challenged with saps from wild type PVX-infected N. benthamiana plants by mechanical inoculation. The cross protection efficacy were evaluated by symptom development on the plants and the virus accumulations were determined by Western blotting assay using antisera against PVX CP. The same cross protection assays were conducted on N. tabacum and tomato plants.
Genetic stability assay
The mutants including E1001A and dM were tested through successive passages to investigate the stability of both the mutations and the mild symptoms. N. benthamiana plants were first infiltrated with Agrobacterium cells carrying the attenuated mutants, and then the sap of leaves of systemic infection were collected to inoculate healthy N. benthamiana plants. The mutants were transferred every 10 days, successively transferred for 5 generations. Primers specific for PVX RdRp were used to clone the RdRp gene from the fifth generation of inoculated N. benthamiana plants.
Northern blotting analysis
The accumulation levels of plus- and minus- strand RNAs in N. benthamiana plants inoculated with wild type and mutant PVX were analyzed by Northern blotting hybridization at 10 dpai. Total RNAs were extracted from systemic leaves using Transzol reagent (Transgen). Five μg of total RNAs was separated by 1.5% agarose formaldehyde gels and transferred to positively charged nylon membranes. The membrane was hybridized overnight at 65 °C with digoxigenin (DIG)-labelled RNA probes (DIG RNA Labelling Kit, Roche Molecular Biochemicals), followed by incubation with anti-DIG Fab fragments. The signal was detected using CDP-star as described in the DIG application manual. The minus- and plus-strand RNA specific probes synthesized separately by PCR amplification of PVX CP followed by in vitro transcription with T7 RNA polymerase. The upstream primer for minus-sense probe was 5′-TAA TAC GAC TCA CTA TAG GGA TGT CGT CAT CAG CTA GCA C-3′ which included nt 5649–5668 of the PVX genome and a T7 promoter (in italics). The downstream primer was 5′-ATG TAG ACG TAG TTA TGG TG-3′ which was complementary to nt 6374–6355 of the PVX genome. The upstream primer for the plus-sense probe was 5′-ATG TCG TCA TCA GCT AGC AC-3′ (identical to 5649–5668 nt in PVX genome), and the downstream primer 5′-TAA TAC GAC TCA CTA TAG GGA TGT AGA CGT AGT TAT GGT G-3′ was complementary to 6374–6355 nt in PVX genome, with the sequence in T7 promoter in italics.
To distinguish the small interfering RNAs (siRNAs) and RNAs of mild mutants from that of the wild type PVX, the saps of plants inoculated with gfp-carrying plasmid pCaPVX440GFP were used for challenging inoculation. Hence, the accumulation levels of siRNA and RNA from the wild type PVX can be determined using probes specific for the gfp gene. Ten days after pre-inoculation with attenuated mutants, the N. benthamiana plants were mechanically inoculated with saps from pCaPVX440GFP infected plants. Ten days later, total RNAs were extracted from the systemic leaves of challenged plants using Transzol reagent (Transgen) and analyzed for viral RNA accumulation by Northern blotting hybridization. The probes used to detect viral RNA were PCR-amplified gfp gene followed by in vitro transcription (Roche). The upstream primer for PCR amplification was 5′-GCG GTA CCA TGA GTA AAG GAG AAG AAC-3′. The downstream primer was 5′-TAA TAC GAC TCA CTA TAG GGT TTG TAG AGC TCA TCC ATG C-3′ which included a T7 promoter (in italics). The same plants were analyzed for the siRNA accumulation in response to virus inoculation using RNAiso for Small RNA kit (Takara). Ten μg of small RNAs were separated on a 15% TBE urea acrylamide gel and subjected to Northern blotting hybridization with probe described above. The membrane was hybridized overnight at 37 °C. The experiment was repeated three times independently.
Results
Effects of mutations in RdRp on virulence, CP and RNA accumulations of PVX
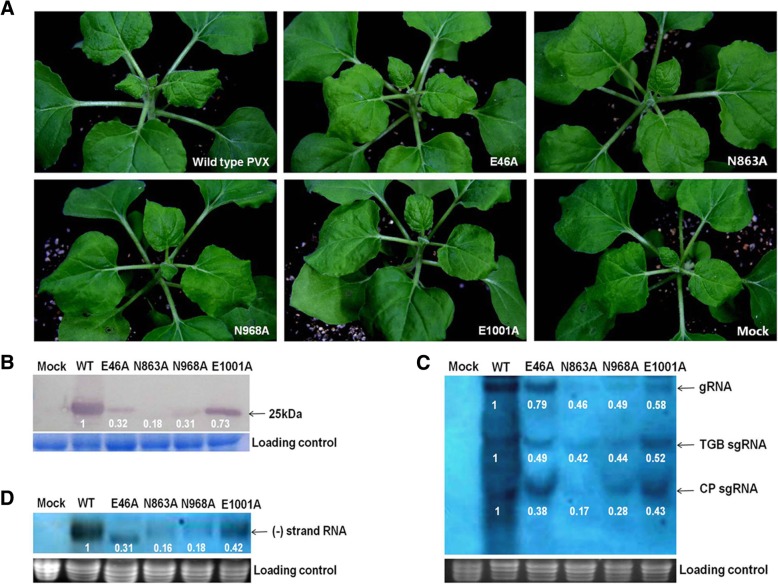
In total, we obtained 40 PVX mutants with amino acid substitution in RdRp (Table 1). Most of these 40 mutants induced severe mosaic symptoms in N. benthamiana plants similar to that the wild type PVX, however, mutants E46A (Glu46 of RdRp mutated to Ala), N863A (Asn863 to Ala), N968A (Asn968 to Ala) and E1001A (Glu1001 to Ala) only induced inconspicuous symptoms on the leaves of inoculated N. benthamiana plants (Fig. 1a), indicating that mutations of Glu46, Asn863, Asn968 and Glu1001 in RdRp significantly reduce the virulence of PVX. The RT-PCR assay revealed that a 506 bp band specific to PVX CP was detected from the upper non-inoculated leaves of plants inoculated with any of these four mutants, showing that these four mutants could infect N. benthamiana plants systemically (data not shown).
Fig. 1.
Symptoms and accumulation levels of wild type (WT) PVX and four attenuated mutants in N. benthamiana plants. a Symptoms of WT PVX and mutants E46A, N863A, N968A and E1001A at 8 days post agro-inoculation (dpai). Mock: Plants inoculated with Argobacterium cells carrying the empty vector pCAMBIA0390. b The CP accumulation level in the systemically infected N. benthamiana leaves at 8 dpai was determined by Western blotting. The relative CP accumulation level of each mutant was marked. c and d The accumulation levels of plus- (c) and minus-strand (d) RNA in N. benthamiana plants infected with WT PVX and four mutants at 10 dpai. The numbers in the figure represented the accumulation levels of each mutant relative to that of WT PVX. (+) strand RNA: PVX genomic RNA; TGB sgRNA: subgenomic RNA for triple gene block; CP sgRNA: subgenomic RNA for coat protein gene; (−) strand RNA: RNA complementary to PVX genomic RNA
In Western blotting assay, the CP accumulation levels of these four mutants in the systemic leaves of N. benthamiana plants were significantly lower than that of wild type PVX (Fig. 1b). The plus- and minus-strand RNA accumulation levels of PVX and the attenuated mutants in N. benthamiana plants were measured by Northern blotting hybridization. The results showed that the levels of plus-strand genomic RNA, TGB and CP sgRNAs in these mutants were lower than that of wild type PVX, and N863A had the lowest RNA accumulation level (Fig. 1c). The minus-strand genomic RNA accumulation levels of these four mutants in N. benthamiana plants were also lower than that of wild type PVX (Fig. 1d).
Taken together, these results indicated that mutations of Glu46, Asn863, Asn968 and Glu1001 in RdRp to Ala had significant impact on the virulence, the CP and RNA accumulation levels of PVX in the systemic leaves of N. benthamiana plants.
Capacity of the attenuated mutants to confer protection against severe PVX infection
When the interval between protective and challenging inoculations was five days, plants pre-inoculated with the attenuated mutants E46A, N863A, N968A, E1001A or the empty vector pCAMBIA0390 showed severe mosaic symptom at 15 days after the challenge inoculation, indicating that an interval of five days is not enough for the attenuated mutants to elicit cross protection (data not shown).
When the protective interval was increased to ten days, all the plants pre-inoculated with E1001A displayed no symptom and a low concentration of PVX CP was detected at 25 days after the challenging inoculation (Fig. 2a and b), indicating that E1001A could provide complete protection against severe PVX infection with an interval of ten days. Plants protected with E46A were absent of viral symptoms at 15 days post challenging inoculation, however, they showed severe mosaic symptoms and high concentration of PVX CP at 25 days post challenging inoculation (Fig. 2a and b), indicating that E46A could delay the infection of wild type PVX. All the plants pre-inoculated with N863A and N968A displayed obvious mosaic symptoms at 15 days after the challenge inoculation and high concentration of PVX CP were detected by Western blotting at 25 days after challenging inoculation (Fig. 2a and b), indicating that an interval of ten days is also not sufficient to elicit cross protection.
Fig. 2.
Cross protection of N. benthamiana plants against the severe infection with wild type PVX. a The protection effects of mutants E46A, N863A, N968A, E1001A and dM against wild type PVX at an interval of 10 days. The symptoms were photographed at 25 days after the challenging inoculation. Plants pre-inoculated with empty plasmid pCAMBIA0390 were used as control. The experiments were repeated three times independently (same below). b The accumulation of wild type PVX CP in E46A-, N863A-, N968A-, E1001A- and dM-protected systemically infected N. benthamiana leaves. The samples were detected by Western blotting using antiserum against PVX CP at 25 days after the challenging inoculation. Total RNAs were used as loading control
When the interval was increased to 15 days, all the N. benthamiana plants pre-inoculated with N863A and N968A showed distinct mosaic symptoms at 15 days post challenge inoculation. In total 12 of 18 N. benthamiana plants pre-inoculated with E46A showed viral symptoms. In contrast, all N. benthamiana plants pre-inoculated with E1001A showed no symptoms at 25 days post challenge inoculation (Table 2). These results indicate that mutant E46A could provide incomplete protection to PVX infection, while E1001A provide complete protection to PVX infection with a protective interval of 15 days. Similar results were obtained on the N. tabacum plants (Table 2).
Table 2.
The protection efficacy of attenuated mutants against severe infection of PVX in N. benthamiana and N. tabacum plants
| Mutants | N. benthamiana | N. tabacum | ||
|---|---|---|---|---|
| 5 days a | 10 days | 15 days | 15 days | |
| E46A | 18/18b | 18/18 | 12/18 | 13/18 |
| N863A | 18/18 | 18/18 | 18/18 | 18/18 |
| N968A | 18/18 | 18/18 | 18/18 | 18/18 |
| E1001A | 18/18 | 0/18 | 0/18 | 0/18 |
| dM | 18/18 | 18/18 | 13/18 | 14/18 |
| Control c | 18/18 | 18/18 | 18/18 | 18/18 |
aProtective intervals of 5, 10 and 15 days were tested for N. benthamiana plants, but only interval of 15 days was tested for N. tabacum plants
bNumber of infected/protected plants. The infection was confirmed with Western blotting assay
cControl: The plants were pre-inoculated with empty vector pCAMBIA0390
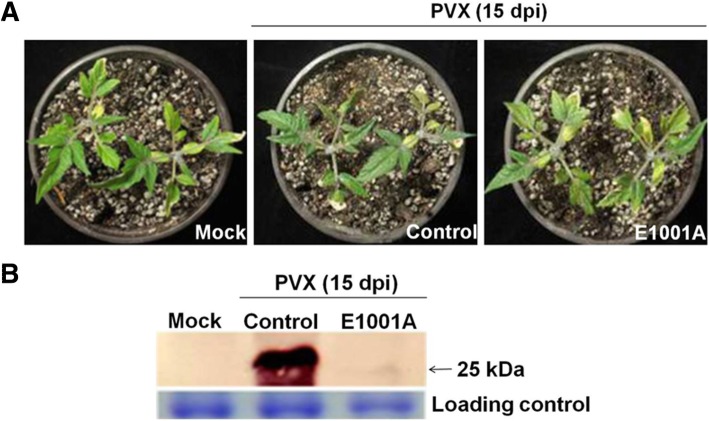
Protection efficacy of E1001A in tomato plants
The above results showed that mutant E1001A provided better cross protection than any other attenuated mutants in N. benthamiana and N. tabacum plants. To examine whether E1001A was able to confer the cross protection against severe infection in other host plants, tomato plants cv. Micro-Tom were first inoculated with E1001A, and then challenge inoculated with wild type PVX at 10 dpai. No symptom was observed and only low concentration of PVX CP were detected by Western blotting at 15 days after challenging inoculation (Fig. 3a and b). Even at 25 days after challenging inoculation, all the plants pre-inoculated with E1001A still showed no symptoms (data not shown). The results showed that E1001A could provide complete protection against severe infection in tomato plants.
Fig. 3.
Cross protection of tomato plants with the E1001A against the challenge inoculation with PVX. a Tomato cv Micro-Tom plants pre- inoculated with empty vector pCAMBIA0390 (control) and E1001A, respectively, were challenged with PVX at 10 dpai. The symptoms were photographed at 15 days after the challenging inoculation. b Virus accumulation in the systemically infected tomato leaves were determined by Western blotting using antiserum against PVX CP at 15 days after the challenging inoculation. Total proteins were used as loading control
Protection effect of a mutant with double mutations to severe infection
To reduce the risk of reverse mutation, we constructed a new PVX mutant designated as dM (i.e. double mutations) in which both Glu46 and Glu1001 of RdRp were mutated to Ala. The mutant dM was still asymptomatic in N. benthamiana plants at 20 dpai (Fig. 4a), and accumulated CP to a level higher than that of E46A but lower than that of E1001A (Fig. 4b). Then we tested its ability to mediate protection against challenging with the wild type PVX. When the protective interval was ten days, all N. benthamiana plants pre-inoculated with dM showed severe symptoms on upper leaves and accumulated high level of CP at 25 days after challenging inoculation (Fig. 2a and b). But when the protective interval was increased to 15 days, of the 18 plants pre-inoculated with dM, only 13 N. benthamiana and 14 N. tabacum plants were systemically infected with the challenging virus (Table 2). These results indicated that mutant dM could provide incomplete protection against severe strain with a protective interval of 15 days.
Fig. 4.
Symptom (a) and virus accumulation (b) of mutant dM with double substitutions in N. benthamiana plants at 20 dpai. Total proteins were used as loading control
Genetic stability of attenuated mutants
The stability test results showed that, after successive transfer of 5 generations, neither of the N. benthamiana plants inoculated with mutants E1001A and dM showed obvious PVX symptoms. The sequencing results indicated that neither mutant E1001A produced recovery mutation after five successive passages in N. benthamiana plants, suggesting that the mutations at Glu46 and E1001 were stable genetically.
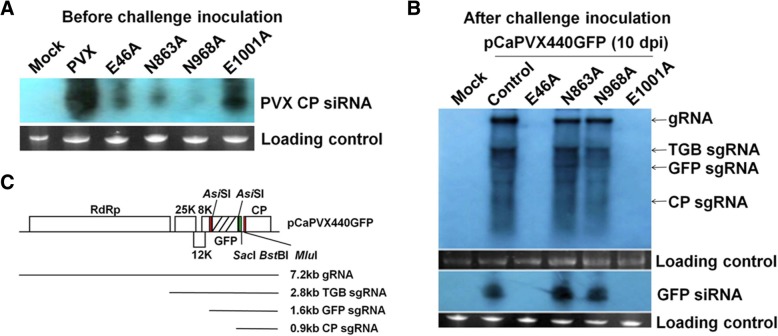
Relationship between siRNA accumulation levels of attenuated mutants and efficacy of cross protection
To explore the relationship between siRNA accumulation levels of attenuated mutants and efficacy of cross protection, we used the attenuated mutants in the protective inoculation and pCaPVX440GFP in the challenging inoculation, and then detect the siRNA with different probes.
The accumulation levels of siRNA from different attenuated mutants were determined via Northern blotting assay using probe specific for the PVX CP gene. E1001A, which provided complete protection to the challenging virus, accumulated the highest level of siRNA, while mutant E46A which conferred incomplete protection against severe infection accumulated low levels of siRNAs. The mutants N863A and N968A which did not show any protection accumulated lower levels of siRNAs (Fig. 5a). These results indicated there existed a positive relationship between the siRNA accumulation levels and the levels of resistance mediated by the attenuated mutants.
Fig. 5.
Accumulation of viral RNA and siRNA in N. benthamiana plants before and after challenging inoculation. a Accumulation of siRNA derived from WT PVX, mutants E46A, N863A, N968A, E1001A at 10 dpai. b Accumulation of virus-derived RNA and siRNA after challenge inoculation with pCaPVX440GFP in the same plants. The protective interval was 10 days. The RNA and siRNA were detected by Northern blotting hybridization at 10 days after challenge with saps from pCaPVX440GFP infected plants. c Genomic organization and approximate sizes of genomic and subgenomic RNAs derived from pCaPVX440GFP. The red rectangles indicated the PVX CP subgenomic promoter (SGP), green rectangle indicated the position of TMV CP SGP. The experiments were repeated three times independently. Total siRNAs were used as loading control
When the protective interval was 10 days, high levels of viral RNAs accumulated in plants pre-inoculated with control plasmid, N863A and N968A, respectively, at ten days after challenging inoculation; however, no viral RNA was detected in plants pre-inoculated with E46A and E1001A (Fig. 5b). The 21–22 nt siRNAs specific for gfp gene were detected in N. benthamiana plants pre-inoculated with control, N863A and N968A, but not in plants pre-inoculated with E46A or E1001A (Fig. 5b), indicating that the invading virus derived from pCaPVX440GFP could not accumulate to detectable levels.
Discussion
In this study we showed that the mutation of Glu46, Asn863, Asn968 or Glu1001 in RdRp to Ala reduced the virulence of PVX and that mutant E1001A displayed the highest cross protection efficacy against severe PVX infection in both N. benthamiana and tomato plants.
The aa residue at position 1422 of the RdRp was reported to be responsible for the symptom development of PVX in Nicotiana plants [53]. In this investigation, for the first time, we demonstrated that four aa, i.e. Glu46, Asn863, Asn968 and Glu1001 of RdRp are novel genetic determinants regulating the PVX virulence, and substitutions with alanine at any of these four residues significantly reduced the symptom severity of PVX (Fig. 1a). Mutations of negatively-charged residues Glu at position Glu46 and Glu1001 to non-polar Ala attenuated the symptoms of PVX. The mutation may change the polarity and/or conformation of RdRp, thus affecting its interaction with potential host factor(s) or other viral products. In other hand, the net charge of RdRp may be critical to maintain its normal function. Similar results were observed in the CP of ZYMV [54]. Furthermore, the substitution of basic Arg180 in helper component-proteinase with hydrophobic aa Ile resulted in mild symptoms in squash [24]. Both amino acids Asn863 and Asn968 of PVX RdRp are postulated glycosylation sites. The substitution of either Asn863 or Asn968 with Ala alleviated the symptom of inoculated plants, implying glycosylation at these two sites in RdRp is critical for PVX virulence. However, more work should be conducted to show if glycosylation occurs in vivo.
The symptom severity of some viruses was positively correlated with viral accumulation level in the infected leaves [45, 46], while that of other viruses was not [42, 44, 55]. The four PVX mutants induced similar mild symptoms but accumulated to different concentrations in the systemic leaves of inoculated host plant (Fig. 1a and b). The differences in virus concentration were positively related to their accumulation levels of viral genomic RNA (Fig. 1b and c). Further analysis indicated that the virus concentrations were positively related to the accumulation levels of plus- and minus- strand RNAs of PVX (Fig. 1b, c and d). It is hypothesized that mutations at these four aa reduced the binding of RdRp with minus strand RNA, thus reducing the accumulation of plus strand RNA, by which they influence the concentration of viral particles.
Cross protection can be separated into three stages: initiation, resistance, and maintenance [24]. It requires an interval between the inoculations of the protector and challenge virus, to allow the full establishment of protector virus in the plant [24, 56]. When the interval was 5 days, none of the four mutants could induce protection to PVX, because the systemic infection has not fully established. When the protective interval was increased to 10 days, E1001A reached a threshold; i.e., a level of extensive viral spread and accumulation sufficient to protect plant from virulent PVX infection. When the interval was 15 days, E1001A provided complete protection, while E46A provided incomplete protection to PVX. These results suggested that longer protective intervals could increase the cross protection efficacy of attenuated mutants against PVX.
The mechanisms underlying cross protection are not fully understood [56, 57]. The two major models for cross protection are CP mediation and RNA silencing mediation [56, 58]. The CP-mediated cross protection model is based on the finding that transgenic plants expressing TMV-CP or plants infected with PVX vector expressing TMV-CP are resistant to TMV infection, suggesting that the uncoating of the second strain was prevented [59, 60]. However, this hypothesis cannot explain the phenomena that CP-defective viruses and viroids can also confer cross protection. Then the RNA silencing model was suggested and accepted to explain the cross protection phenomenon for RNA and DNA viruses, as well as for viroids [56]. Plants pre-inoculated with E1001A and E46A displayed complete and incomplete resistance, respectively. The accumulation levels of siRNA were positively correlated with the efficiency of resistance (Fig. 5a and b). Plants inoculated with E46A had similar CP accumulation with that of N968A, however, E46A had much higher genomic and siRNA accumulations than that of N968A, which could explain why E46A had higher resistance efficiency than N968A (Fig. 1b and c; Fig. 5a and b). However, the mechanism underlying cross protection remains to be elucidated.
Based on results on a potyvirus Turnip mosaic virus, Kung et al. [57] proposed four criteria for the identification of a useful mild strain for effective cross-protection. Our results agree with their ideas in that the mild strain should produce stable mild symptoms and yet should not cause severe adverse effects on the protected host, and especially, a moderate and sustained titer of a protective virus is necessary to trigger cross-protection. The mutant E1001A accumulated highest level of CP and conferred highest cross protection efficacy. However, in this paper, we just introduced mutations to the RdRp of PVX, without changing the suppression activity of P25.
Among the methods available for mild strain screening, site-directed mutagenesis has unique advantages. Once an infectious clone of certain virus is constructed, it is easy to produce large number of mutants and evaluate their potential in cross protection. The data in this study and previous reports [16, 24] illustrated that the strategy we adopted here offers a rapid and effective way to create mild strains that have potential in cross protection. By modifying available mild strains of Papaya ringspot virus, the effectiveness of HA5–1 against heterologous isolates was improved [14]. By using mutagenesis, Kung et al. generated mild strain Tu-GK which provided complete cross-protection against Turnip mosaic virus in N. benthamiana and Arabidopsis plants [57].
The major risk of cross protection is that mild strains may become virulent ones by reverse mutations [3, 56]. One solution to reduce such risk is to create mild strains with two or more mutations. The mutant dM, which contains two mutations at both Glu46 and Glu1001, was asymptomatic and accumulated to a level lower than that of E1001A. It can provide protection against severe PVX infection in N. benthamiana and N. tabacum plants when the interval was 15 days. Genetic stability test showed that the both mutants E1001A and E 46A were stable genetically and did not show obvious symptom after successive transfer for five generations.
Cross protection is a promising strategy for biological control of plant viral diseases. The elucidation of viral virulence determinants via reverse genetics is an important step towards screening mild strains for cross protection. The findings of this research further confirmed the potential and validity of genetic engineering for producing mild strains and laid a solid foundation for the control of PVX via cross protection.
Conclusions
The result in this study showed that substitutions of RdRp Glu46, Asn863, Asn968 or Glu1001 to Ala drastically attenuated PVX symptom and mutants E1001A and E46A could protect Nicotiana benthamiana and tomato from severe infection. Mutant dM which carries mutations in both Glu46 and Glu1001 provided effective resistance to wild type PVX. Both mutants E1001A and E46A had no recovery mutation after 5 successive transfers with interval of 10 days. However, further studies will be needed to test if the mutants will keep stable genetically after transferring for longer times, and to dissect the molecular mechanisms regulating cross protection. The findings of this research laid a solid foundation for the control of PVX via cross protection.
Acknowledgements
The authors would like to acknowledge the support from Shandong ‘Double Top’ Program (SYL2017XTTD11) and ‘Taishan Scholar’ Construction Project (TS201712023). The authors are grateful to Dr. Beth Stokes in Mississippi State University for the careful language edition.
Funding
This work was supported by grants awarded to X. Li from the National Natural Science Foundation of China (NSFC, 31571895, 31720103912) and Shandong Provincial Natural Science Foundation (HN201703, KN238–201602).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- aa
Amino acids
- CP
Coat protein
- DIG
Digoxigenin
- M-MLV
Moloney Murine Leukemia Virus
- NC
Nitrocellulose
- ORF
Open reading frame
- PVX
Potato virus X
- RdRp
RNA-dependent RNA polymerase
- RT-PCR
Reverse transcription-polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- siRNAs
Small interfering RNAs
- TBST
Tris-buffered saline containing 0.05% Tween-20
- TGB
Triple gene block
- TMV
Tobacco mosaic virus
Authors’ contributions
Designed the study: QC, XL and YT; performed the experiments and analyzed the data: QC, YW, JL; wrote the paper: QC, YL, YT and XL; revised the manuscript critically for important intellectual content: QC, ZG and JY. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Q. Q. Cong, Email: cong.qianqian@163.com
Y. Wang, Email: wangying@lyu.edu.cn
J. Liu, Email: liujincc612@126.com
Y. F. Lan, Email: xiaolan407@163.com
Z. K. Guo, Email: guozhaokui66@163.com
J. G. Yang, Email: jinguangyang@caas.cn
X.-D. Li, Phone: 86 538 8242523, Email: xdongli@sdau.edu.cn
Y. P. Tian, Phone: 86 538 8242523, Email: yanping.tian@sdau.edu.cn
References
- 1.Fulton RW. Practices and precautions in the use of cross protection for plant virus disease control. Annu Rev Phytopathol. 1986;24:67–81. doi: 10.1146/annurev.py.24.090186.000435. [DOI] [Google Scholar]
- 2.Kurihara Y, Watanabe Y. Cross-protection in Arabidopsis against crucifer tobamovirus cg by an attenuated strain of the virus. Mol Plant Pathol. 2003;4:259–269. doi: 10.1046/j.1364-3703.2003.00174.x. [DOI] [PubMed] [Google Scholar]
- 3.Sherwood JL. Mechanisms of cross-protection between plant virus strains. Plant resistance to viruses. Wiley, Chichester. CIBA Found Symp. 1987;133:136–150. [Google Scholar]
- 4.McKinney HH. Mosaic diseases in the Canary Islands, West Africa and Gibraltar. J Agric Res. 1929;39:557–578. [Google Scholar]
- 5.Nakazono-Nagaoka E, Takahashi T, Shimizu T, Kosaka Y, Natsuaki T, Omura T, et al. Cross-protection against Bean yellow mosaic virus (BYMV) and Clover yellow vein virus by attenuated BYMV isolate M11. Phytopathology. 2009;99:251–257. doi: 10.1094/PHYTO-99-3-0251. [DOI] [PubMed] [Google Scholar]
- 6.Costa AS, Muller GW. Tristeza control by cross protection: a US-Brazil cooperative success. Plant Dis. 1980;64:538–541. doi: 10.1094/PD-64-538. [DOI] [Google Scholar]
- 7.Ieki H, Yamaguchi A, Kano T, Koizumi M, Iwanami T. Control of stem pitting disease caused by citrus tristeza virus using protective mild strains in navel orange. Ann Phytopathol Soc Jpn. 1997;63:170–175. doi: 10.3186/jjphytopath.63.170. [DOI] [Google Scholar]
- 8.Motoyoshi F, Nishiguchi M. Control of virus diseases by attenuated virus strains, comparison between attenuated strains of cucumber green mottle mosaic virus and tobacco mosaic virus. Gamma Field Symposia. 1988;27:91–109. [Google Scholar]
- 9.Kosaka Y, Fukunishi T. Multiple inoculation with three attenuated viruses for the control of cucumber virus disease. Plant Dis. 1997;81:733–738. doi: 10.1094/PDIS.1997.81.7.733. [DOI] [PubMed] [Google Scholar]
- 10.Sayama H, Sato T, Kominato M, Natsuaki T, Kaper JM. Field testing of a satellite-containing attenuated strain of cucumber mosaic virus for tomato protection in Japan. Phytopathology. 1993;83:405–410. doi: 10.1094/Phyto-83-405. [DOI] [Google Scholar]
- 11.Tien P, Wu GS. Satellite RNA for the biocontrol of plant disease. Adv Virus Res. 1991;39:321–339. doi: 10.1016/S0065-3527(08)60799-X. [DOI] [PubMed] [Google Scholar]
- 12.Owor B, Legg JP, Okao-Okuja G, Obonyo R, Kyamanywa S, Ogenga-Latigo MW. Field studies of cross protection with cassava mosaic geminiviruses in Uganda. J Phytopathol. 2004;152:243–249. doi: 10.1111/j.1439-0434.2004.00837.x. [DOI] [Google Scholar]
- 13.Yeh SD, Gonsalves D. Evaluation of induced mutants of Papaya ringspot virus for control by cross protection. Phytopathology. 1984;74:1086–1091. doi: 10.1094/Phyto-74-1086. [DOI] [Google Scholar]
- 14.You BJ, Chiang CH, Chen LF, Su WC, Yeh SD. Engineered mild strains of Papaya ringspot virus for broader cross protection in cucurbits. Phytopathology. 2005;95:533–540. doi: 10.1094/PHYTO-95-0533. [DOI] [PubMed] [Google Scholar]
- 15.Agüero J, Gómez-Aix C, Sempere RN, García-Villalba J, García-Núñez J, Hernando Y, et al. Stable and broad spectrum cross-protection against Pepino mosaic virus attained by mixed infection. Front Plant Sci. 2018;9:1810. doi: 10.3389/fpls.2018.01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon JY, Ahn HI, Kim M, Tsuda S, Ryu KH. Pepper mild mottle virus pathogenicity determinants and cross protection effect of attenuated mutants in pepper. Virus Res. 2006;118:23–30. doi: 10.1016/j.virusres.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Kosaka Y, Fukunishi T. Attenuated isolates of soybean mosaic virus derived at a low temperature. Plant Dis. 1993;77:882–886. doi: 10.1094/PD-77-0882. [DOI] [Google Scholar]
- 18.Burgyan J, Gaborjanyi R. Cross-protection and multiplication of mild and severe strains of TMV in tomato plants. J Phytopathol. 1984;110:156–167. doi: 10.1111/j.1439-0434.1984.tb03404.x. [DOI] [Google Scholar]
- 19.Goto T, Nemoto M. Studies on control of plant virus diseases by interference of attenuated virus-selection of TMV attenuated strain and influence on various plants inoculated with the attenuated strain. Res Bull Hokkaido Natl Agric Exp Sta. 1971;99:67–76. [Google Scholar]
- 20.Oshima N. Control of tomato mosaic disease by attenuated virus. Jpn Agric Res Q. 1981;14:222–228. [Google Scholar]
- 21.Rast ATB. Variability of tobacco mosaic virus in relation to control of tomato mosaic in glasshouse tomato crops by resistance breeding and cross protection. Agric Res Rep. 1975;834:1–76. [Google Scholar]
- 22.Yang G, Qiu BS, Liu XG, Li Y, Wang XF. Nonsense mutations of replicase and movement protein genes contribute to the attenuation of an avirulent tomato mosaic virus. Virus Res. 2002;87:119–128. doi: 10.1016/S0168-1702(02)00025-4. [DOI] [PubMed] [Google Scholar]
- 23.Kosaka Y, Ryang BS, Kobori T, Shiomi H, Yasuhara H, Kataoka M. Effectiveness of an attenuated Zucchini yellow mosaic virus isolate for cross-protecting cucumber. Plant Dis. 2006;90:67–72. doi: 10.1094/PD-90-0067. [DOI] [PubMed] [Google Scholar]
- 24.Lin SS, Wu HW, Jan FJ, Hou RF, Yeh SD. Modifications of the helper component-protease of Zucchini yellow mosaic virus for generation of attenuated mutants for cross protection against severe infection. Phytopathology. 2007;97:287–296. doi: 10.1094/PHYTO-97-3-0287. [DOI] [PubMed] [Google Scholar]
- 25.Bercks, R. 1970. Potato virus X. Commonwealth Mycological Institute (Kew, England)/Association of Applied Biologists Descriptions of Plant Viruses, no. 4.
- 26.Huisman MJ, Linthorst HJ, Bol JF, Cornelissen JC. The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J Gen Virol. 1988;69:1789–1798. doi: 10.1099/0022-1317-69-8-1789. [DOI] [PubMed] [Google Scholar]
- 27.Skryabin KG, Kraev AS, Morozov SY, Rozanov MN, Chernov BK, Lukasheva LI, et al. The nucleotide sequence of potato virus X RNA. Nucleic Acids Res. 1988;16:10929–10930. doi: 10.1093/nar/16.22.10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu XQ, Jia JL, Zhang CL, Li XD, Wang YJ. Phylogenetic analyses of an isolate obtained from potato in 1985 revealed potato virus X was introduced to China via multiple events. Virus Genes. 2010;40:447–451. doi: 10.1007/s11262-010-0468-5. [DOI] [PubMed] [Google Scholar]
- 29.Yu XQ, Wang HY, Lan YF, Zhu XP, Li XD, Fan ZF, et al. Complete genome sequence of a Chinese isolate of Potato virus X and analysis of genetic diversity. J Phytopathol. 2008;156:346–351. doi: 10.1111/j.1439-0434.2007.01365.x. [DOI] [Google Scholar]
- 30.Forster RL, Bevan MW, Harbison SA, Gardner RC. The complete nucleotide sequence of the potexvirus white clover mosaic virus. Nucleic Acids Res. 1988;16:291–303. doi: 10.1093/nar/16.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angell SM, Davies C, Baulcombe DC. Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology. 1996;216:197–201. doi: 10.1006/viro.1996.0046. [DOI] [PubMed] [Google Scholar]
- 32.Beck DL, Guilford PJ, Voot DM, Andersen MT, Forster RLS. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology. 1991;183:695–702. doi: 10.1016/0042-6822(91)90998-Q. [DOI] [PubMed] [Google Scholar]
- 33.Morozov SY, Zakhariev VM, Chernov BK, Prasolov VS, Kozlov YV, Atabekov JG, et al. The analysis of the primary structure and localization of the coat protein gene on the genomic RNA of potato virus X. Dokl Akad Nauk SSSR. 1983;271:211–215. [Google Scholar]
- 34.Cockerham G. Proceedings of the second conference on potato virus diseases. 1955. pp. 89–92. [Google Scholar]
- 35.Moreira A, Jones RAC, Fribourg CE. Properties of a resistance- breaking strain of potato virus X. Ann Appl Biol. 1980;95:93–103. doi: 10.1111/j.1744-7348.1980.tb03974.x. [DOI] [Google Scholar]
- 36.Querci M, Baulcombe D, Goldbach R, Salazar L. Analysis of the resistance-breaking determinants of potato virus X (PVX) strain HB on different potato genotypes expressing extreme resistance to PVX. Phytopathology. 1995;85:1003–1010. doi: 10.1094/Phyto-85-1003. [DOI] [Google Scholar]
- 37.Tozzini A, Ceriani M, Cramer P, Palva E, Hopp H. PVX MS, a new strain of potato virus that overcomes the extreme resistance gene Rx. J Phytopathol. 1994;141:241–248. doi: 10.1111/j.1439-0434.1994.tb01467.x. [DOI] [Google Scholar]
- 38.Salaman RN. Protective inoculation against a plant virus. Nature. 1933;131:468. doi: 10.1038/131468a0. [DOI] [Google Scholar]
- 39.Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Csorba T, Pantaleo V, Burgyán J. RNA silencing: an antiviral mechanism. Adv Virus Res. 2009;75:35–230. doi: 10.1016/S0065-3527(09)07502-2. [DOI] [PubMed] [Google Scholar]
- 41.Gal-On A. A point mutation in the FRNK motif of the potyvirus helper component-protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology. 2000;90:467–473. doi: 10.1094/PHYTO.2000.90.5.467. [DOI] [PubMed] [Google Scholar]
- 42.Saenz P, Quiot L, Quiot JB, Candresse T, Garcia JA. Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Mol Plant-Microbe Interact. 2001;14:278–287. doi: 10.1094/MPMI.2001.14.3.278. [DOI] [PubMed] [Google Scholar]
- 43.Wu HW, Lin SS, Chen KC, Yeh SD, Chua NH. Discriminating mutations of HC-pro of Zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol Plant-Microbe Interact. 2010;23:17–28. doi: 10.1094/MPMI-23-1-0017. [DOI] [PubMed] [Google Scholar]
- 44.Diveki Z, Salanki K, Balazs E. The necrotic pathotype of the Cucumber mosaic virus (CMV) ns strain is solely determined by amino acid 461 of the 1a protein. Mol Plant-Microbe Interact. 2004;17:837–845. doi: 10.1094/MPMI.2004.17.8.837. [DOI] [PubMed] [Google Scholar]
- 45.Hagiwara K, Ichiki TU, Ogawa Y, Omura T, Tsuda S. A single amino acid substitution in 126-kDa protein of Pepper mild mottle virus associates with symptom attenuation in pepper; the complete nucleotide sequence of an attenuated strain, C-1421. Arch Virol. 2002;147:833–840. doi: 10.1007/s007050200030. [DOI] [PubMed] [Google Scholar]
- 46.Lewandowski DJ, Dawson WO. A single amino acid change in Tobacco mosaic virus replicase prevents symptom production. Mol Plant-Microbe Interact. 1993;6:157–160. doi: 10.1094/MPMI-6-157. [DOI] [PubMed] [Google Scholar]
- 47.Tsuda S, Kubota K, Kanda A, Ohki T, Meshi T. Pathogenicity of Pepper mild mottle virus is controlled by the RNA silencing suppression activity of its replication protein but not the viral accumulation. Phytopathology. 2007;97:412–420. doi: 10.1094/PHYTO-97-4-0412. [DOI] [PubMed] [Google Scholar]
- 48.Ziebell H, Payne T, Berry JO, Walsh JA, Carr JP. A cucumber mosaic virus mutant lacking the 2b counter-defence protein gene provides protection against wild-type strains. J Gen Virol. 2007;88:2862–2871. doi: 10.1099/vir.0.83138-0. [DOI] [PubMed] [Google Scholar]
- 49.Voinnet O, Lederer C, Baulcombe DC. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/S0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 50.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu HT, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Cong QQ, Lan YF, Geng C, Li X, Liang YC, et al. Development of new potato virus X-based vectors for gene over- expression and gene silencing assay. Virus Res. 2014;191:62–69. doi: 10.1016/j.virusres.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Kagiwada S, Yamaji Y, Komatsu K, Takahashi S, Mori T, Hirata H, et al. A single amino acid residue of RNA-dependent RNA polymerase in the Potato virus X genome determines the symptoms in Nicotiana plants. Virus Res. 2005;110:177–182. doi: 10.1016/j.virusres.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Kimalov B, Gal-On A, Stav R, Belausov E, Arazi T. Maintenance of coat protein N-terminal net charge and not primary sequence is essential for zucchini yellow mosaic virus systemic infectivety. J Gen Virol. 2004;85:3421–3430. doi: 10.1099/vir.0.80417-0. [DOI] [PubMed] [Google Scholar]
- 55.Johansen IE, Dougherty WG, Keller KE, Wang D, Hampton RO. Multiple viral determinants affect seed transmission of pea seedborne mosaic virus in Pisum sativum. J Gen Virol. 1996;77:3149–3154. doi: 10.1099/0022-1317-77-12-3149. [DOI] [PubMed] [Google Scholar]
- 56.Gal-On A, Shiboleth YM. Cross-protection. In: Loebenstein G, Carr JP, editors. Natural resistance mechanism of plants to viruses. Berlin: Springer; 2006. pp. 261–288. [Google Scholar]
- 57.Kung Y, Lin P, Yeh S, Hong S, Chua N, Liu L, et al. Genetic analyses of the FRNK motif function of Turnip mosaic virus uncover multiple and potentially interactive pathways of cross- protection. Mol Plant-Microbe Interact. 2014;27:944–955. doi: 10.1094/MPMI-04-14-0116-R. [DOI] [PubMed] [Google Scholar]
- 58.Baulcombe DC. Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beachy RN. Coat-protein-mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philos Trans R Soc Lond Ser B Biol Sci. 1999;354:659–664. doi: 10.1098/rstb.1999.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu B, Stubbs G, Culver JN. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology. 1998;248:188–198. doi: 10.1006/viro.1998.9280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.