Figure 3.
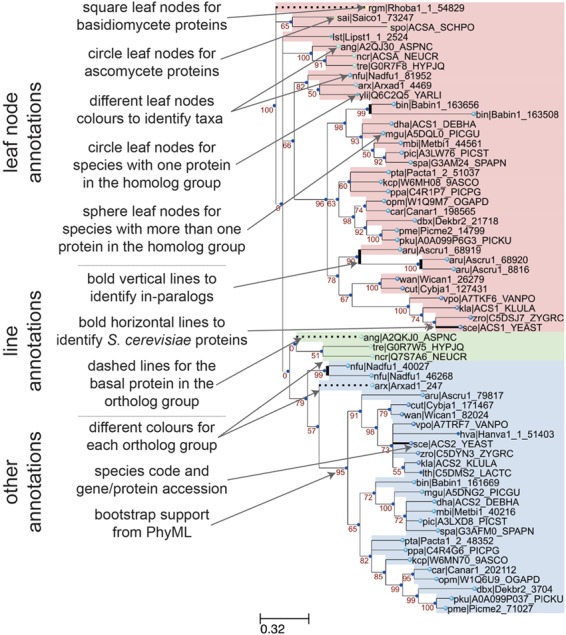
Annotation features of a sample phylogenetic tree in AYbRAH. Square and circle leaves indicate protein sequences in Basidiomycota or Ascomycota, respectively. Leaf nodes are colored based on taxonomic groups. Circle leaves are used for proteins with no paralogs in the same species, whereas sphere leaves are used to designate proteins with paralogs in the same species. Vertical bold lines indicate species-lineage expansions, which are sometimes called in-paralogs or co-orthologs (61). Horizontal bold lines designate S. cerevisiae proteins, which is the most widely studied eukaryote. Dashed lines indicate the most anciently diverged protein sequence in the ortholog group. Ortholog groups can be identified by color groups to help the visual inspection of ortholog assignments. The leaf names include a three-letter species code and a sequence accession. Internal nodes are labeled with the bootstrap values from phylogenetic reconstruction with PhyML.
