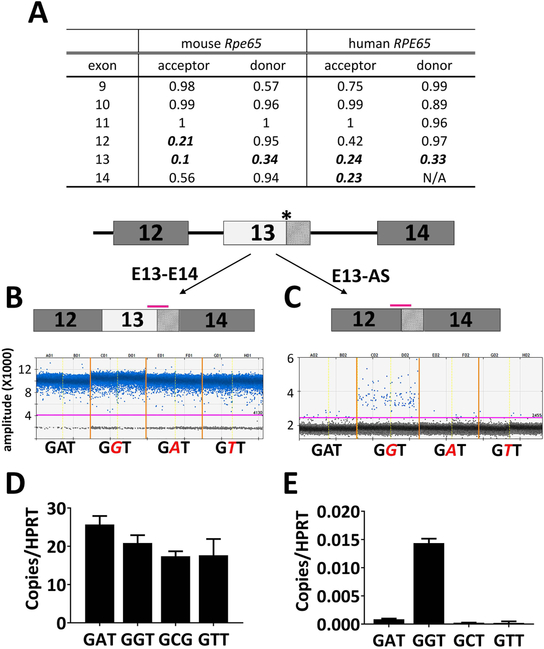
Figure 9.
Defects in splicing are readily detected in human RPE65 minigene carrying the c.1430G mutation. (A) Scores of splicing signals strength for Exons 9–14 of WT RPE65 in mouse and human. High score indicates a strong splicing signal with the default cut-off score for an effective splicing site being 0.4. Note that in both mouse and human, Exon 13 has a weak splicing strength as the scores are below 0.4 and are shown in italic bold fonts. (B-E) minigene constructs that include the human genomic sequence of either WT (c.1430A) or mutants (c.1430G, c.1430C, c.1430T) were transfected into HEK293T cells and the transcribed variants were analyzed via ddPCR. One dimensional ddPCR plots for quantification of the regular spliced read-through transcript E13-E14 (B) , and the alternatively spliced variant E13-AS (C) that lacks the first 91 nucleotide of Exon 13. Data were derived from duplicates of construct transfections. Each point represents a single droplet, which is scored as positive (blue colored) or negative (grey) depending on the fluorescent amplitude. The expression levels were normalized to the expression of reference gene HPRT1 (D and E). While the read-through transcript E13-E14 was readily detected in both the cells expressing WT and the mutant minigenes (B and D), the alternative spliced E13-AS was only enriched in the cells expressing mutant minigene (C and E). The asterisk marks the c.1430A>G mutation. Pink bars denote positions of the Taqman probes for mRNA level quantification.