Abstract
New approaches are needed for understanding and treating acute myeloid leukemia (AML). MicroRNAs (miRs) are important regulators of gene expression in all cells and disruption of their normal expression can lead to changes in phenotype of a cell, in particular the emergence of a leukemic clone. We collected peripheral blood samples from 10 adult patients with newly diagnosed AML, prior to induction chemotherapy, and 9 controls. Two and a half ml of whole blood was collected in Paxgene RNA tubes. MiRNA was purified using RNeasy mini column (Qiagen). We sequenced approximately 1000 miRs from each of 10 AML patients and 9 controls. In subset analysis, patients with NPM1 and FLT3 mutations showed the greatest number of miRNAs (63) with expression levels that differed from control with adjusted p-value of 0.05 or less. Some of these miRs have been described previously in association with leukemia, but many are new. Our approach of global sequencing of miRs as opposed to microarray analysis removes the bias regarding which miRs to assay and has demonstrated discovery of new associations of miRs with AML. Another strength of our approach is that sequencing miRs is specific for the 5p or 3p strand of the gene, greatly narrowing the proposed target genes to study further. Our study provides new information about the molecular changes that lead to evolution of the leukemic clone and offers new possibilities for monitoring relapse and developing new treatment strategies.
Introduction
One area of research that has grown explosively in the past 15 years is the study of non-coding RNA. MiRNAs are 19–22 nucleotide long non-coding RNAs which regulate the expression of genes by sequence-specific binding to mRNA to either promote or block its translation [1]. This is a powerful level of epigenetic control for gene expression that can influence the phenotype of a cell [2]. Several authors have examined the role of miRNA in the transformation of hematopoietic stem cells into leukemic cells [3–8]. It is now well established that miRNAs play a role in blocking differentiation of leukemic cells and promoting their unchecked cell division [9,10].
To date, researchers have analyzed miRNA expression in leukemic cells using arrays or RT-PCR. Our work is the first quantitative sequencing of miRNAs found in peripheral blood from patients with newly diagnosed acute myeloid leukemia (AML). We demonstrate here proof of principle for this relatively simple method. We compare the expression levels to normal controls to find statistically significant increase or decrease in levels of expression of approximately 1000 miRNAs. Subset analysis of AML patients with FLT3 or NPM1 mutations yields further information regarding statistically significant changes in expression of specific miRNAs.
Materials and methods
This protocol was reviewed and approved by the Upstate Medical University Institutional Review Board. Blood was obtained following written informed consent from patients at University Hospital with newly diagnosed AML, prior to start of induction chemotherapy. Blood was also obtained following written informed consent from healthy volunteers involved in patient care at Upstate. 2.5 ml of whole blood was collected into Paxgene RNA preparation tubes and stored at -80C for batch processing. miRNA was purified using a Qiagen miRNA purification kit. The yield and quality of the RNA samples was assessed using the Agilent Bioanalyzer prior to library construction using the Illumina TruSeq Small RNA Sample Prep protocol (Illumina; San Diego, California). Multiplexed samples of RNA that exceed quality control metrics (RIN > 6.0) were run on an Illumina NextSeq500 instrument at a targeted depth of 10 million reads per sample. After filtering and trimming of index and adapter sequences, whole genome alignment of the miR FASTQ reads was performed using the Homo sapiens/hg21 reference genome in the SHRiMPS aligner included in the miRNAs analysis application available in BaseSpace (Illumina), as well as the sRNA Toolbox application suite.
Statistical method
The analysis of the RNA-seq data was performed following the pipeline available from the limma packages [11] in the Bioconductor project [12]. Log2counts per million (logCPM) transformation was applied before normalization and linear model fitting. Empirical Bayes moderation was carried out to obtain robust estimates of gene-wise variability and the final p-values from the linear model with appropriately designed contrasts were adjusted by the Benjamin–Hochberg procedure for a targeted false discover rate of 0.05. Volcano plots and boxplots were used to graphically examine the differences between groups.
Results
We sequenced all miRNAs in peripheral whole blood from ten patients with newly diagnosed AML and nine normal controls. Table 1 shows the characteristics of the ten patients who entered the study. There were five males and five females, ages ranged from 42 to 87. Initial white blood cell (WBC) count ranged from 1.1 x 103/ul to 88 x 103/ul, with blast percentage of 3.8 to 73. Phenotype was determined by standard hematopathology staining for surface markers. For all ten patients, we recorded presence (pos) or absence (neg) of NPM1 or FLT3 mutations and cytogenetic analysis.
Table 1. Characteristics of patients in study.
| Age | Gender | Initial WBC x 1000/ul | Blast % | Phenotype | NPM1 | FLT3 | Cytogenetics |
|---|---|---|---|---|---|---|---|
| 55 | M | 1.5 | 73 | Acute biphenotypic T/myeloid | neg | pos | normal |
| 42 | F | 9 | 17 | Erythroleukemia from CML | neg | neg | complex |
| 55 | F | 29 | 13 | Acute monocytic leukemia | pos | pos | normal |
| 87 | F | 88 | 18 | Acute myelomonocytic leukemia | pos | pos | normal |
| 57 | M | 10.9 | 35 | Acute myeloid leukemia | pos | pos | trisomy 8 |
| 72 | M | 28 | 4.5 | Acute myeloid leukemia | pos | neg | normal |
| 59 | M | 1.8 | 4 | Acute erythroid leukemia | neg | neg | normal |
| 72 | M | 30 | 23 | Acute myeloid leukemia | neg | neg | normal |
| 87 | F | 2.3 | 42 | Acute myeloid leukemia | neg | neg | normal |
| 49 | F | 1.1 | 3.8 | Acute promyelocytic leukemia | neg | neg | 15:17 |
We obtained sequence for 996 miRNAs and averaged the amount of each miRNA for the ten AML patients and the nine controls. Levels of expression were compared with a t-test and the adjusted P-value was calculated for each miRNA. In our first analysis of all ten patients versus controls, we identified only thirteen miRNAs with expression levels that showed a statistically significant difference (adjusted P-value less than 0.05) [13].
We then performed subset analysis of our data and found that patients who were double negative for mutations (NPM1- and FLT3-) had only one miRNA with adjusted P-value less than 0.05, while patients who were double positive for mutations (NPM1+ and FLT3+) had 63 miRNAs with adjusted P-value less than 0.05 [14]. Results are shown in Fig 1 (volcano plots).
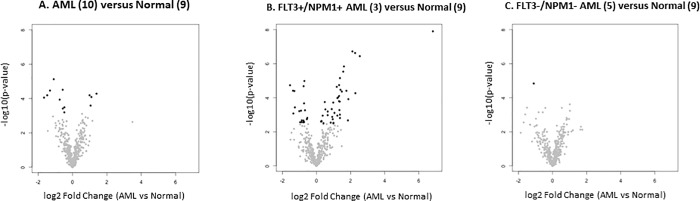
Fig 1. Volcano plots showing differences in miRNAs expressed in leukemia versus control peripheral blood.
X-axis: log2 Fold Change Y-axis: -log10(p-value). A. AML versus Normal (n = 10) B. FLT3+/NPM1+ AML versus Normal (n = 3) C. FLT3-/NPM1- AML versus Normal (n = 5).
In Fig 1, the volcano plot for the comparison of all ten AML patients versus control is shown in panel A. Log2 Fold Change is shown on the x-axis and–log10 adjusted p-value is shown on the y-axis. Points in dark font indicate miRNAs with statistically significant log fold change and adjusted p-value (four are increased in AML, nine are decreased in AML). Panel B shows the volcano plot for the subset of patients who were double positive (NPM1+/FLT3+, n = 3), with 63 miRNAs having statistically significant change in expression. Similarly, panel C shows the volcano plot for the subset of AML patients who were double negative (NPM1- /FLT3-, n = 5) with only one statistically significant change in miRNA expression. Clearly, the subset analysis identified a greater number of miRNAs with statistically significant change in expression in the AML patients with NPM1 and FLT3 mutations (double positive).
Table 2 shows the first twenty (out of 996) miRNAs that were identified, listed by rank order of adjusted P-value, lowest to highest, in comparison of AML versus control samples. The center list is from all ten AML patients versus nine controls. The first 13 show statistically significant difference with adjusted p-value less than 0.05. The list on the left is from the comparison of double negative patients (NPM1-/FLT3-) (n = 5) to nine controls. Only hsa-miR-328-3p shows a statistically significant difference with adjusted p-value less than 0.05. Interestingly, some of the other miRNAs in the top twenty do show a statistically significant change in the double positive patients (identified by color bars). The list on the right is from the comparison of double positive patients (NPM1+/FLT3+) (n = 3) to nine controls. There are 63 (out of 996) miRNAs with statistically significant difference in expression and adjusted p-value less than 0.05, but only the top twenty (lowest adjusted p-values) are shown here. The color bars are used to demonstrate which miRNAs are found in two out of the three, or all three samples sets.
Table 2. Differential expression of miRNAs in patients versus controls ranked by adjusted P-value.
| NPM1- FLT3- (n = 5) | adj.P.Val | AML (N = 10) | adj.P.Val | NPM1+ FLT3+ (n = 3) | adj.P.Val |
|---|---|---|---|---|---|
| hsa-miR-328-3p | 0.014 | hsa-miR-328-3p | 0.007 | hsa-miR-10a-5p | 1.24E-05 |
| hsa-miR-24-3p | 0.078 | hsa-miR-106b-3p | 0.011 | hsa-miR-146b-5p | 7.79E-05 |
| hsa-let-7i-5p | 0.078 | hsa-let-7i-5p | 0.011 | hsa-miR-181a-3p | 7.79E-05 |
| hsa-miR-34a-5p | 0.078 | hsa-miR-181a-3p | 0.011 | hsa-miR-155-5p | 8.95E-05 |
| hsa-miR-1229-3p | 0.078 | hsa-miR-409-3p | 0.011 | hsa-miR-199b-5p | 0.0002 |
| hsa-miR-181a-3p | 0.089 | hsa-miR-10b-5p | 0.011 | hsa-miR-24-3p | 0.0004 |
| hsa-miR-3200-5p | 0.089 | hsa-miR-24-3p | 0.011 | hsa-miR-19b-3p | 0.0009 |
| hsa-miR-106b-3p | 0.089 | hsa-miR-126-5p | 0.011 | hsa-miR-425-5p | 0.0012 |
| hsa-miR-15a-5p | 0.104 | hsa-miR-3200-5p | 0.013 | hsa-let-7a-3p | 0.0018 |
| hsa-miR-744-5p | 0.135 | hsa-miR-23a-3p | 0.026 | hsa-miR-4301 | 0.0018 |
| hsa-let-7d-5p | 0.135 | hsa-miR-323b-3p | 0.029 | hsa-miR-181a-2-3p | 0.0018 |
| hsa-miR-23a-3p | 0.135 | hsa-miR-652-3p | 0.032 | hsa-miR-335-5p | 0.0018 |
| hsa-miR-625-5p | 0.135 | hsa-miR-181a-2-3p | 0.049 | hsa-miR-10b-5p | 0.0024 |
| hsa-miR-10b-5p | 0.142 | hsa-miR-22-3p | 0.056 | hsa-miR-328-3p | 0.0024 |
| hsa-miR-126-5p | 0.186 | hsa-miR-3960 | 0.074 | hsa-miR-142-3p | 0.0024 |
| hsa-miR-15b-3p | 0.193 | hsa-miR-625-5p | 0.077 | hsa-miR-3615 | 0.0024 |
| hsa-miR-125b-2-3p | 0.193 | hsa-miR-181b-5p | 0.08 | hsa-miR-23a-3p | 0.0025 |
| hsa-miR-409-3p | 0.193 | hsa-miR-4772-3p | 0.087 | hsa-miR-27a-3p | 0.0029 |
| hsa-miR-3960 | 0.193 | hsa-miR-744-5p | 0.087 | hsa-miR-181b-5p | 0.0041 |
| has-miR-4301 | 0.193 | has-miR-146b-5p | 0.087 | has-miR-125b-2-3p | 0.0047 |
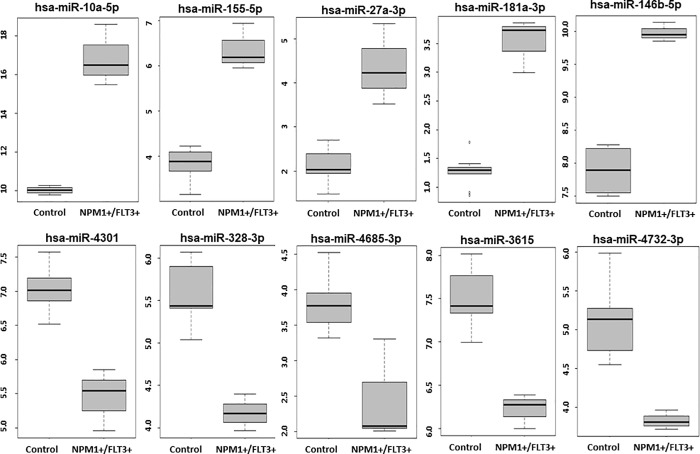
Fig 2 shows boxplots of the relative expression levels for the ten miRNAs showing greatest increase or greatest decrease when comparing NPM1+/FLT3+ AML to controls. The boxplots show the average expression level (log2 counts per million) for each of ten miRNAs in controls versus NPM1+/FLT3+AML blood samples.
Fig 2. Boxplots showing relative expressions levels of ten miRNAs with statistically significant difference between control and AML NPM1+/FLT3+ patient samples.
Table 3 shows all 63 miRNAs that had statistically significant differences in expression between NPM1+/FLT3+ patients and controls, listed in decreasing order of log2Fold Change (log2FC). Positive values of log2FC indicate increased expression in the AML patients, while negative values of log2FC indicate decreased expression in the AML patients. MiRs that have not previously been associated with AML are indicated with asterisks.
Table 3. Differential expression of miRNAs between NPM1+/FLT3+ AML patients and controls.
| miRNA | Log2FC | miRNA | Log2FC |
|---|---|---|---|
| hsa-miR-10a-5p | 6.830569383 | hsa-miR-339-5p | 0.654592192 |
| hsa-miR-155-5p | 2.540153625 | hsa-miR-378c | 0.652454498 |
| hsa-miR-27a-3p | 2.282394076 | hsa-miR-371b-5p | 0.521138094 |
| hsa-miR-181a-3p | 2.26616154 | hsa-miR-500a-3p * | 0.481850139 |
| hsa-miR-146b-5p | 2.107437782 | hsa-miR-378d | 0.407412804 |
| hsa-miR-21-3p | 1.870878887 | hsa-miR-181b-3p | 0.360659064 |
| hsa-miR-340-5p | 1.844418662 | hsa-miR-1910-5p * | 0.295654388 |
| hsa-miR-142-3p | 1.755215629 | hsa-miR-195-5p | 0.287016156 |
| hsa-miR-199b-5p | 1.608767963 | hsa-miR-6882-5p * | 0.286345176 |
| hsa-miR-24-3p | 1.567277012 | hsa-miR-3667-5p * | 0.285098138 |
| hsa-miR-23a-3p | 1.484152009 | hsa-miR-3922-3p * | 0.279184469 |
| hsa-miR-10b-5p | 1.406513049 | hsa-miR-548al * | 0.268147495 |
| hsa-miR-6503-3p * | 1.374880947 | hsa-miR-199a-5p | -0.542007567 |
| hsa-miR-19b-3p | 1.373298073 | hsa-miR-361-3p | -0.564971933 |
| hsa-miR-28-5p * | 1.370999597 | hsa-miR-106b-3p | -0.677684789 |
| hsa-miR-10a-3p | 1.359816952 | hsa-miR-425-5p * | -0.711429318 |
| hsa-miR-582-3p * | 1.343371672 | hsa-miR-532-3p * | -0.726695535 |
| hsa-let-7a-3p | 1.325948775 | hsa-miR-92a-3p | -0.730022499 |
| hsa-miR-181b-5p | 1.325353217 | hsa-miR-181a-2-3p | -0.758535453 |
| hsa-miR-30e-3p | 1.322709817 | hsa-miR-6511b-3p * | -0.768444175 |
| hsa-miR-125b-2-3p | 1.258254645 | hsa-miR-574-3p * | -0.782318226 |
| hsa-miR-29b-3p | 1.224950228 | hsa-miR-3605-3p * | -0.866437008 |
| hsa-miR-29a-3p | 1.223591909 | hsa-miR-3940-3p * | -0.886685286 |
| hsa-miR-335-5p | 1.184936831 | hsa-miR-3200-5p* | -0.911067568 |
| hsa-miR-3688-3p * | 1.054040212 | hsa-miR-484 * | -0.961659564 |
| hsa-miR-222-3p | 0.996724055 | hsa-miR-486-5p | -1.010732451 |
| hsa-miR-769-5p * | 0.986880376 | hsa-miR-4732-3p * | -1.270122308 |
| hsa-miR-451a | 0.981368744 | hsa-miR-3615 * | -1.299778948 |
| hsa-miR-1307-5p * | 0.94986408 | hsa-miR-4685-3p * | -1.35782737 |
| hsa-miR-338-3p | 0.918728355 | hsa-miR-328-3p | -1.380320316 |
| hsa-miR-223-3p | 0.891956558 | hsa-miR-4301 * | -1.554755355 |
| hsa-miR-23a-5p | 0.830519732 |
*indicates miRNAs not previously associated with leukemia
Discussion
Rather than analyze by microarray which would identify only 300–400 miRNAs, we chose to sequence all miRNAs that were obtained in whole blood samples from patients with newly diagnosed AML and controls. We quantified expression of 996 miRNAs from each patient and control and performed statistical analyses to determine which miRNAs were increased or decreased in AML patients versus normal controls. Subset analysis revealed the most differences in patients with double positive AML (NPM1+/FLT3+ mutations). NPM1 mutations are the most common genetic abnormalities in AML (50–60% of cytogenetically normal AML and 30% of all AML) [15]. Up to one-third of NPM1+ patients also have a mutation in FLT3, which counteracts the favorable prognosis of the NPM1 mutation [16]. The NPM1 gene encodes a 32-kDA protein that is involved in numerous cellular processes. It can function as an oncogene and a tumor suppressor gene depending on expression levels, interacting proteins, and cellular compartmentalization. Dozens of mutations have been described in NPM1 and how they contribute to leukemogenesis is not known. Finding 63 distinct miRNAs either upregulated or downregulated in NPM1+ /FLT3+ AML suggests that there may be multiple molecular pathways disrupted that contribute to the leukemic phenotype.
On our list of miRNAs associated with NPM1+/FLT3+ AML, miR-10a-5p showed the most statistically significant adjusted p-value as well as the highest fold change. This miRNA has been described in patients with NPM1 mutations and high expression levels are associated with good response to induction chemotherapy [17]. More recent studies demonstrate a role for miR-10a/b in regulating the proliferation and differentiation of HL-60 leukemic cells in vitro [18].
One miR family that we identified with statistically significant change was the miR-181 family (181a-3p, 181a-2-3p and 181b-5p). This family has been shown by others to be consistently increased in AML patients [8,19,20]. There are numerous possible target mRNAs for the miR-181 family and it has been proposed as a therapeutic target.
Another report has shown that miR-199b is consistently decreased in AML and we also found that to be the case in our patients [21]. We found increased expression of miR-223-3p and this has been shown to regulate granulopoiesis in mice [22].
We compared our list of miRNA differences determined by Nextgen sequencing to lists of miRNA differences found by hybridization arrays or RT-PCR [10, 23–27]. Many of the miRNAs identified by these methods were the same as ours, confirming our ability to generate meaningful data with our approach. Differences in miRNAs identified can be attributed to differences in technique of sample processing (whole blood versus isolated cells or serum). Identification of novel miRNAs in association with AML are likely due to the greater sensitivity of our approach, i.e. sequencing all miRNAs obtained in whole blood.
New therapeutic approaches are being created to target specific miRNAs in leukemic patients [28,29]. As we continue to investigate the role of miRNAs in leukemogenesis, the approaches of diagnosis, treatment, and post-treatment monitoring will be greatly improved.
Supporting information
This table shows the statistical analysis for each of 996 miRNAs for patients with NPM1+/FLT3+ AML versus normal controls. Results are listed in decreasing order of adjusted P. Value.
(XLSX)
Acknowledgments
Thanks to Katherine Gilligan for assistance with formatting.
Data Availability
All relevant data are within the manuscript, Supporting Information files, and on the NCBI GEO repository with accession number: GSE128079 (https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE128079)
Funding Statement
The authors received no specific funding for this work.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D, Boldin MP, O'connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008;9(8):839–845. 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol 2008. Jul;15(4):352–358. [DOI] [PubMed] [Google Scholar]
- 4.Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget 2016;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A 2008. October 7;105(40):15535–15540. 10.1073/pnas.0808266105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isken F, Steffen B, Merk S, Dugas M, Markus B, Tidow N, et al. Identification of acute myeloid leukaemia associated microRNA expression patterns. Br J Haematol 2008;140(2):153–161. 10.1111/j.1365-2141.2007.06915.x [DOI] [PubMed] [Google Scholar]
- 7.Cammarata G, Augugliaro L, Salemi D, Agueli C, Rosa ML, Dagnino L, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol 2010;85(5):331–339. 10.1002/ajh.21667 [DOI] [PubMed] [Google Scholar]
- 8.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood 2011. January 27;117(4):1121–1129. 10.1182/blood-2010-09-191312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood 2017. September 14;130(11):1290–1301. 10.1182/blood-2016-10-697698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trino S, Lamorte D, Caivano A, Laurenzana I, Tagliaferri D, Falco G, et al. MicroRNAs as New Biomarkers for Diagnosis and Prognosis, and as Potential Therapeutic Targets in Acute Myeloid Leukemia. Int J Mol Sci 2018. February 3;19(2): 10.3390/ijms19020460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015. April 20;43(7):e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 2005. August 15;21(16):3439–3440. 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- 13.Pandita A, Basnet A, Ramadas P, Poudel A, Anand A, Saad N, et al. Micro RNAs in Acute Myeloid Leukemia 2016. Blood 2016;128:5252 [Google Scholar]
- 14.Gilligan DM, Pandita A, Ramadas P, Poudel A, Saad N, Anand A, Basnet A, Middleton. MicroRNA profiling in AML. Cancer Research 2018(78):5413. [Google Scholar]
- 15.Ziai JM, Siddon AJ. Pathology Consultation on Gene Mutations in Acute Myeloid Leukemia. Am J Clin Pathol 2015;144(4):539–554. 10.1309/AJCP77ZFPUQGYGWY [DOI] [PubMed] [Google Scholar]
- 16.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008;358(18):1909–1918. 10.1056/NEJMoa074306 [DOI] [PubMed] [Google Scholar]
- 17.Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood 2014. April 10;123(15):2412–2415. 10.1182/blood-2013-10-532374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi L, Sun L, Jin Z, Zhang S, Shen Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett 2018. April;15(4):5611–5619. 10.3892/ol.2018.8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng H, Lal K, Yang FF, Chen J. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancer genetics 2015;208(5):225–229. 10.1016/j.cancergen.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su R, Lin H, Zhang X, Yin X, Ning H, Liu B, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene 2015;34(25):3226–3239. 10.1038/onc.2014.274 [DOI] [PubMed] [Google Scholar]
- 21.Favreau AJ, McGlauflin RE, Duarte CW, Sathyanarayana P. miR-199b, a novel tumor suppressor miRNA in acute myeloid leukemia with prognostic implications. Experimental hematology & oncology 2016;5(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trissal MC, DeMoya RA, Schmidt AP, Link DC. MicroRNA-223 Regulates Granulopoiesis but Is Not Required for HSC Maintenance in Mice. PloS one 2015;10(3):e0119304 10.1371/journal.pone.0119304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 2008. May 14;3(5):e2141 10.1371/journal.pone.0002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daschkey S, Rottgers S, Giri A, Bradtke J, Teigler-Schlegel A, Meister G, et al. MicroRNAs distinguish cytogenetic subgroups in pediatric AML and contribute to complex regulatory networks in AML-relevant pathways. PLoS One 2013;8(2):e56334 10.1371/journal.pone.0056334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008. May 15;111(10):5078–5085. 10.1182/blood-2008-01-133355 [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Chen H, Bai J, He A. MicroRNA: an important regulator in acute myeloid leukemia. Cell Biol Int 2017. September;41(9):936–945. 10.1002/cbin.10770 [DOI] [PubMed] [Google Scholar]
- 27.Shivarov V, Bullinger L. Expression profiling of leukemia patients: key lessons and future directions. Exp Hematol 2014. August;42(8):651–660. 10.1016/j.exphem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 28.Velu CS, Chaubey A, Phelan JD, Horman SR, Wunderlich M, Guzman ML, et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J Clin Invest 2014;124(1):222–236. 10.1172/JCI66005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 2015;29(11):2143–2153. 10.1038/leu.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table shows the statistical analysis for each of 996 miRNAs for patients with NPM1+/FLT3+ AML versus normal controls. Results are listed in decreasing order of adjusted P. Value.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript, Supporting Information files, and on the NCBI GEO repository with accession number: GSE128079 (https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE128079)