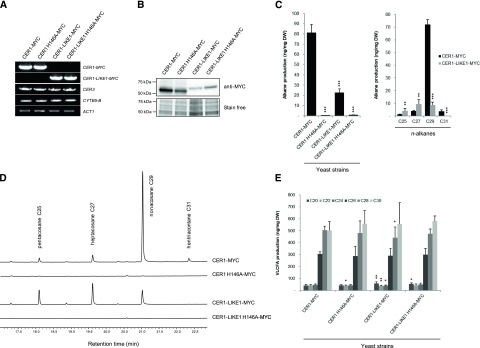
Figure 2.
Functional characterization of CER1-LIKE1. A, Semiquantitative analysis of steady-state CER1-MYC, CER1-LIKE1-MYC, CER3, and CYTB5-B transcripts (identity indicated on the right) in INVSur4# yeast cotransformed with MYC-tagged wild-type or mutated forms of CER1 or CER1-LIKE1 (indicated at top), CER3, and CYTB5-B. The yeast Actin1 (ACT1) gene was used as a constitutively expressed control. B, Immunoblot analysis of protein extracts of the different INVSur4# yeast strains expressing MYC-tagged wild-type or mutated forms of CER1 or CER1-LIKE1. Total proteins were imaged using Stain-free technology and used for equal loading control. C, Mean values of total alkanes and individual alkanes of the different INVSur4# yeast strains reported in ng per mg of dry weight (DW). Error bars represent sd (n = 4). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). D, gas chromatography-mass spectrometry traces of the hydrocarbon fractions after separation of total lipid using TLC of the different INVSur4# yeast strains. E, Mean values of VLC fatty acyl (VLCFA) methyl esters of the different INVSur4# yeast strains reported in ng per mg of dry weight (DW). Error bars represent sd (n = 4). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).