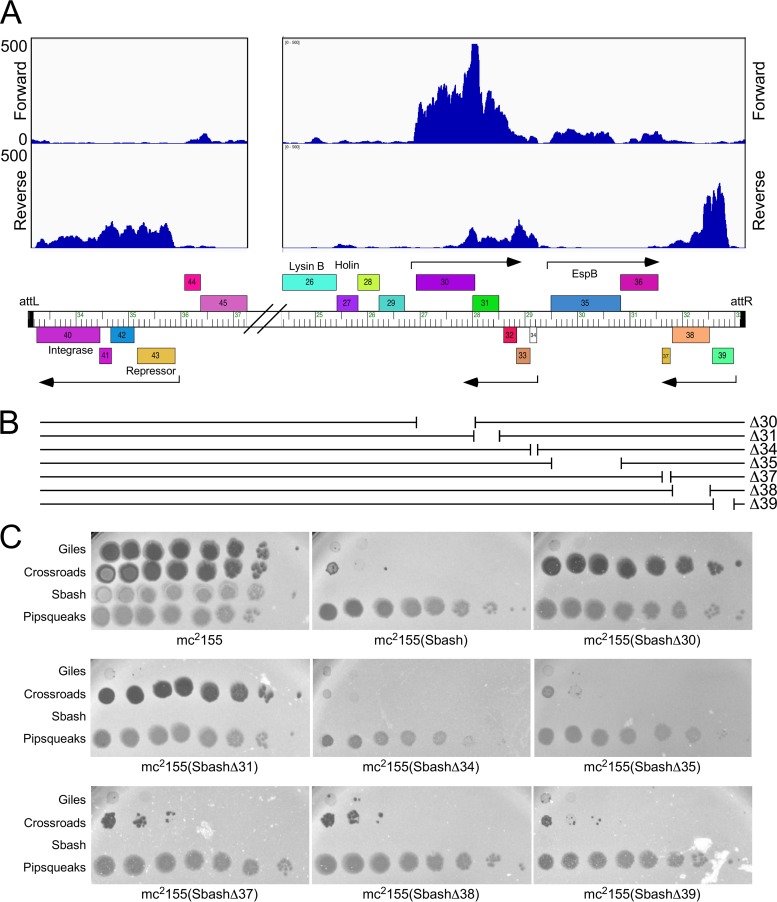
FIG 2.
Sbash genes required for viral defense. (A) Lysogenic expression of Sbash prophage genes. RNAseq reads are mapped to the Sbash prophage, shown in its integrated orientation from attL to attR, with the relevant part of the genome map shown at the bottom. Putative lysogenically expressed operons are represented by arrows. The RNAseq reads are strand specific, and those mapping to forward and reverse DNA orientations are indicated. Results of RNAseq analysis of the whole prophage are shown in Fig. S1. (B) Sbash deletion derivatives. The seven deletion mutants are depicted, showing the region removed in each mutant. Genomes are aligned with the map shown at the top. (C) Plating efficiencies of phages on mutant Sbash lysogens. Ten-fold serial dilutions of phages Giles, Crossroads, Sbash, and Pipsqueaks were plated onto lawns of M. smegmatis mc2155 and mc2155(Sbash) and of mutant derivatives in which individual Sbash genes were knocked out. Sbash 30 and 31 are both required for defense against Crossroads; none of the deleted genes are required for defense against Giles infection.