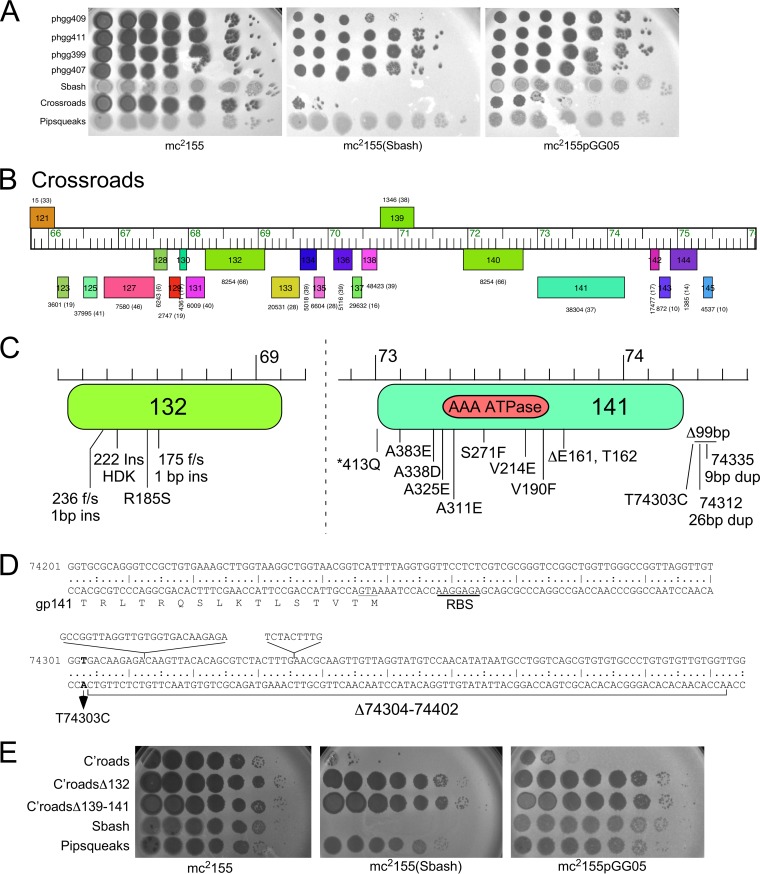
FIG 5.
Characterization of Crossroads defense escape mutants. (A) Plating of Crossroads defense escape mutants (DEMs) on host strains. Crossroads mutants were isolated as escapees on lawns of either a Sbash lysogen or a pGG05 recombinant strain, purified, and shown to escape Sbash prophage defense. Ten-fold serial dilutions of DEMs (phgg409, phgg411, phgg399, phgg407) were plated on bacterial lawns as indicated. (B) Organization at the extreme right end of the Crossroads genome showing genes 121 to 145. The genome is displayed as described for Fig. 1. (C) Mapping of Crossroads DEM mutations. Defense escape mutants of Crossroads were purified and sequenced, and the locations of the mutations were mapped to gene 132 and 141 and the 141 to 142 intergenic region. The location of the AAA ATPase domain in gp141 (residues 186 to 323) predicted by pfam is shown. (D) DEM mapping in the Crossroads 141-to-142 intergenic region. The DNA sequence is shown for the beginning of the leftward-transcribed gene 141 and its amino acid sequence, together with the sequences immediately upstream. The ATG translation start codon for gene 141 is underlined, and the ribosome binding site (RBS) is indicated. Four DEM mutations are shown: a single base substitution at coordinate 74,303, 26-bp and 9-bp duplications, and a 99-bp deletion. (E) Crossroads 132 or 139 to 141 is required for targeting by Sbash 30/31. Serial dilutions of a Crossroads mutant in which gene 132 is deleted (C’roadsΔ132) or genes 139 through 141 are deleted (C’roadsΔ139–141) together with Crossroads (C’roads), Sbash, and Pipsqueaks were plated onto lawns of M. smegmatis mc2155, a Sbash lysogen strain, and a pGG05 recombinant strain.