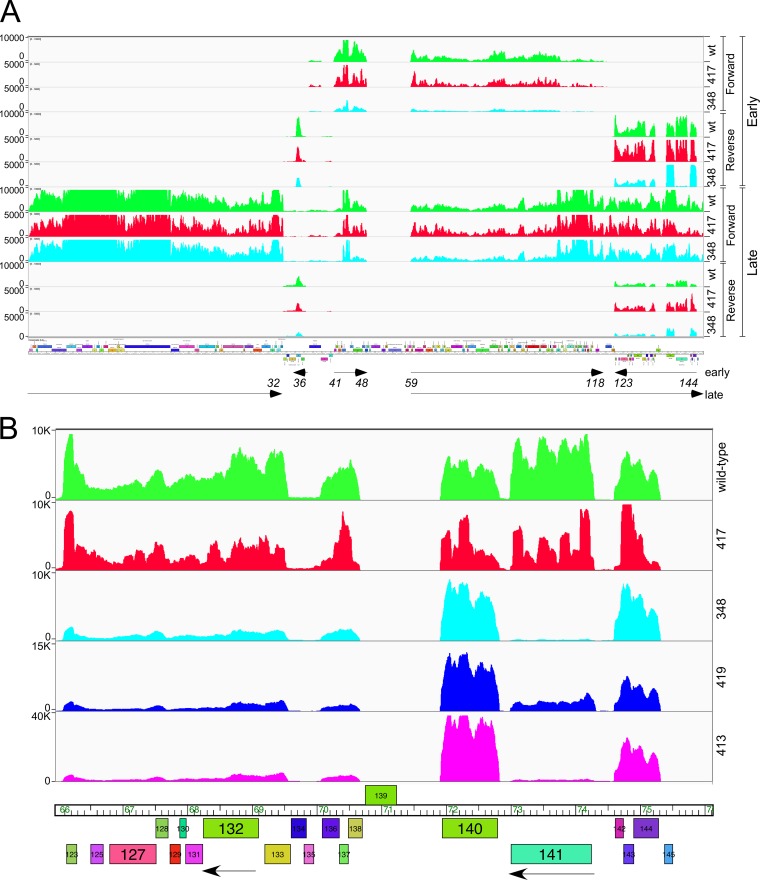
FIG 7.
RNAseq analysis of Crossroads and DEM mutants. (A) RNA was isolated at early (30 min) and late (150 min) time points after infection of M. smegmatis with wild-type Crossroads (green) or DEMs phgg348 (aqua) and phgg417 (red); reads mapping to forward and reverse strands are shown as indicated. Arrows at the bottom indicate the early and late transcribed regions, with gene numbers denoting the positions. (B) Expanded view of RNAseq reads mapping to the reverse strands at the right end of the genome. DEM mutants phgg419 (blue) and phgg413 (magenta) are shown in addition to those shown in panel A and are similarly colored. Arrows indicate leftward transcription of the two genes, 132 and 141, in which DEM mutants mapped. Reads are shown for the reverse strand only.