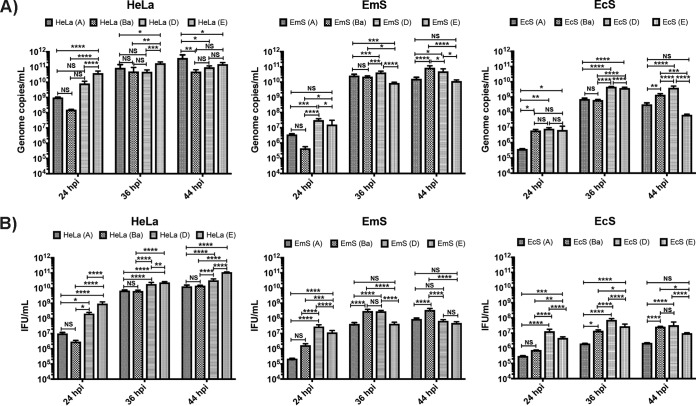
FIG 3.
Infectious progeny production by C. trachomatis strains A/Har-13, Ba/Apache-2, D/UW-3, and E/Bour in HeLa, EcS, and EmS cells. HeLa 229 cells and primary EcS and EmS cells from the same patient were infected with an ocular or urogenital C. trachomatis strain. At 24, 36, and 44 hpi, the cultures were serially diluted onto the same cell types from the same patients as the primary infection. (A) C. trachomatis genome copy numbers by strain and cell type at designated time points. Genomic DNA was extracted from each culture, and qPCR was performed to determine the genome copy number for each reinfectivity assay (see Materials and Methods). (B) Numbers of IFU per milliliter by strain and cell type at designated time points. Numbers of IFU per milliliter were determined for each reinfectivity assay (see Materials and Methods). A one-way ANOVA was performed at each time point (with a Fishers LSD posttest). *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001; NS, not significant. The data reflect three independent experiments for HeLa cells and for primary cells from 3 to 6 patients.