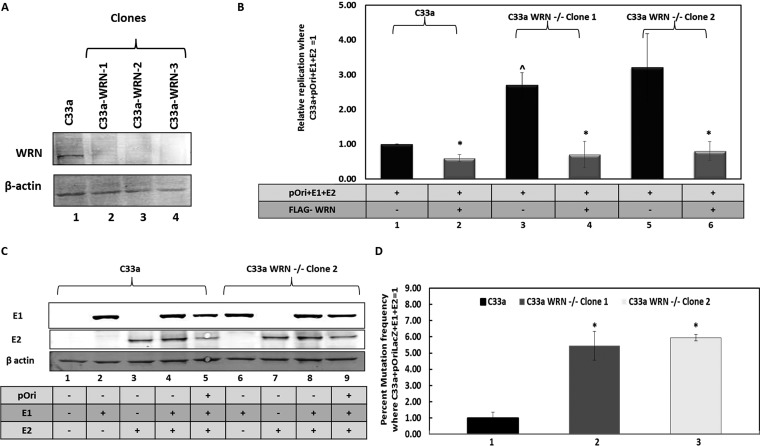
FIG 2.
Deletion of WRN generates a phenotype similar to that after the loss of SIRT1. (A) CRISPR/Cas9 was used to generate clonal cell lines (lanes 2 to 4) that lacked WRN expression. (B) E1-E2 replication levels were elevated in the absence of WRN (compare lanes 3 and 5 with lane 1), while overexpression of wild-type WRN repressed replication (lanes 2, 4, and 6). In the WRN CRISPR cells (lanes 3 to 6), FLAG-WRN overexpression resulted in replication levels similar to those in C33a wild-type cells with FLAG-WRN overexpression (compare lanes 4 and 6 with lane 2). In each cell line, FLAG-WRN resulted in a statistically significant decrease in replication (*; P value was less than 0.05; standard error bars are shown). There is a statistically significant increase in replication levels (^) in C33a WRN–/– clone 1 cells compared with those in C33a wild-type cells (P value was less than 0.05). The histogram depicts the average of results from five independent experiments. (C) The expression levels of E1 and E2 are not affected by the absence of WRN. (D) The absence of WRN results in a significantly enhanced (*) mutation frequency for E1-E2 replication (compare lanes 2 and 3 with lane 1) (P value was less than 0.05; standard error bars are shown). The experiment was repeated three times.