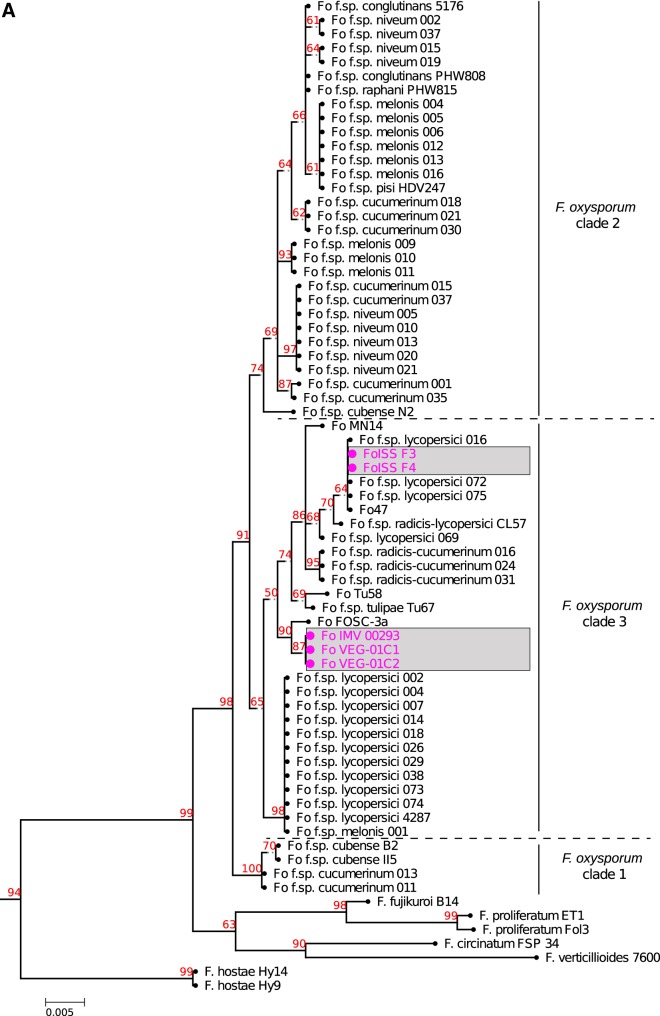
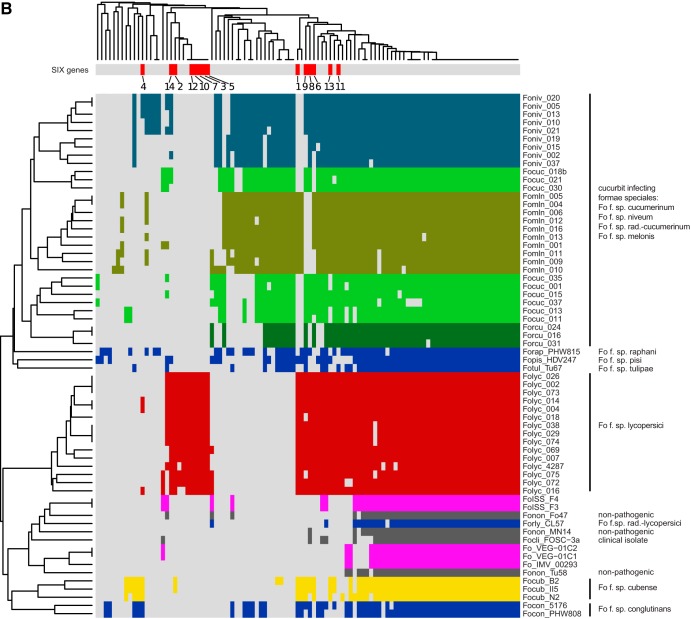
FIG 1.
Phylogenetic relationship of ISS-F3/F4 among other F. oxysporum isolates. (A) The phylogenetic tree constructed from EF-1α sequences of 62 F. oxysporum isolates places ISS-F3/F4 in clade 3, belonging to a clonal lineage shared by lycopersici_016, lycopersici_072, lycopersici_075, and Fo47. Other Fusarium species (bottom) were included to root the tree. The bootstrap confidence (in percent; 1,000 replicates performed) is shown in red numbers in the tree. (B) Presence/absence profiles of ISS-F3/F4 with 62 other strains based on 104 candidate effector proteins. Colored boxes indicate the presence of a particular effector gene, with gray indicating its absence in the genome. The list of effector genes is shown in Data Set S4. Formae speciales that cause disease in the same host cluster together, as seen by the groupings of green (cucurbit-infecting strains), red (tomato-infecting strains), and yellow (banana-infecting strains). Dark gray represents non-plant-pathogenic isolates, and pink represents strains isolated from or flown to the ISS. Blue represents other plant pathogens. ISS-F3/F4 contained relatively few effector genes compared to the number found in the plant pathogens (green, red, yellow, and blue) and showed the most similar pattern (number and type) to Fo47, a non-plant-pathogenic soil isolate with a biocontrol function.