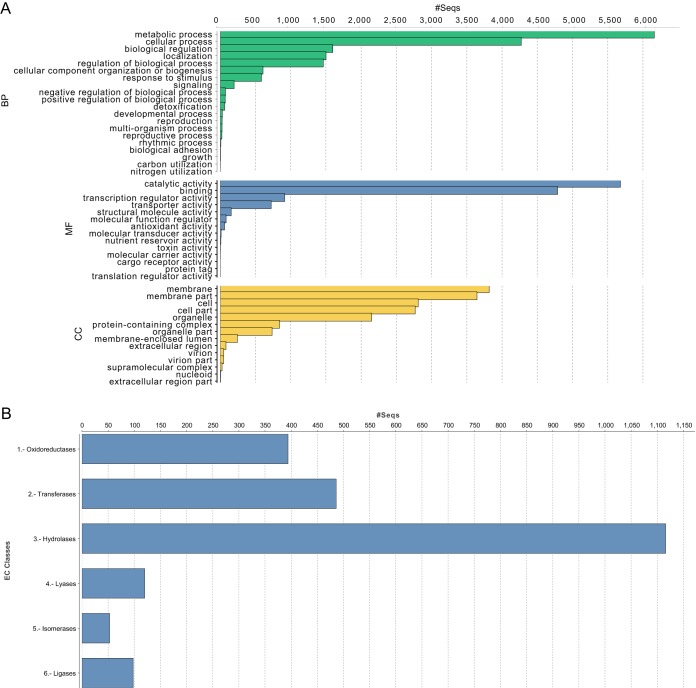
FIG 3.
Summary of gene ontology (GO) annotation of the predicted genes in ISS-F3/F4. Predicted genes in ISS-F3/F4 were predicted using AUGUSTUS and then annotated using Blast2GO. Functions could be assigned to 10,789 of the 16,648 genes in ISS-F3 and 11,305 of the 16,729 genes in ISS-F4. (A) Bar charts summarizing the most abundant GO terms for ISS-F3. BP, biological processes; MF, metabolic function; CC, cellular components. (B) Bar charts summarizing the abundance of enzyme classes in ISS-F3. NB, ISS-F3/F4 had the same distributions of GO and enzyme class abundances, and thus, only ISS-F3 is shown for simplicity. The distributions in ISS-F3/F4 were similar to Fo47 (non-plant-pathogenic soil isolate) and FOSC-3a (clinical isolate) (data not shown).