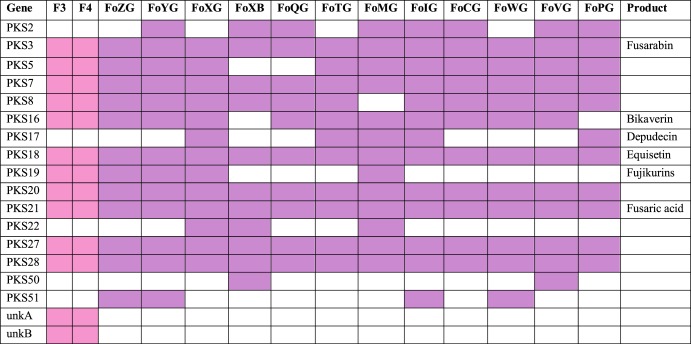
FIG 5.
Presence/absence profile of polyketide synthases (PKSs) detected in the genomes of ISS-F3/F4. The sequences of predicted genes that had a ketoacyl synthase domain (one of 3 essential domains in polyketide synthases [PKSs]) were compared with BLAST against a PKS database (35) to determine which PKSs ISS-F3/F4 had the ability to produce. Eleven PKSs found in ISS-F3/F4 are also present in 12 other F. oxysporum species that have been previously studied (35). However, there were two additional PKSs, unkA and unkB, in ISS-F3/F4 that did not have any matches in the database. The first column represents the PKS, the last column shows the polyketide that the PKS makes (if known), and the middle columns indicate the F. oxysporum strains. Colored boxes indicate the presence of a specific PKS in the genome. FoZG, Fo47; FoYG, FOSC-3a; FoXG, lycopersici 4287; FoXB, Fo5176; FoQG, raphani NRRL 54005; FoTG, vasinfectum NRRL 25433; FoMG, melonis NRRL 26406; FoIG, cubense tropical race 4; FoCG, radicis-lycopersici 26381; FoWG, MN25; FoVG, pisi (HDV 247); FoPG, conglutinans race 2 NRRL 54008. Amino acid sequences for the unknown PKSs in ISS-F3 can be found in Data Set S1 (g14942 and g11694), and those for ISS-F4 can be found in Data Set S2 (g15849 and g12099). NB, the sequences are identical between ISS-F3/F4.