FIGURE 2.
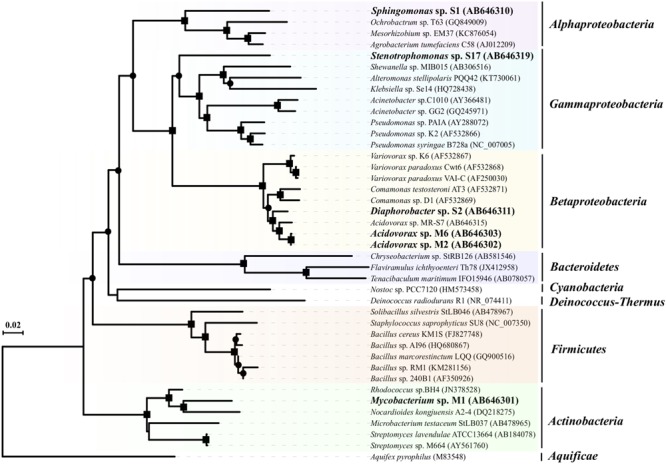
Phylogenetic affiliations of the AHL-degrading isolates obtained in this study and the previously known AHL degraders based on their almost full length 16S rRNA gene sequences. The phylogenetic tree was constructed by neighbor-joining (NJ) method with Kimura’s correction. The 16S rRNA gene sequence of Aquifex pyrophilus (M83548) was used as an outgroup. Bootstrap values of >50% and >80% estimated using neighbor-joining and maximum-likelihood methods (1,000 replications) are shown by circle and square at branching points, respectively. The six new AHL-degrading strains isolated in this study are shown in name with boldface.
