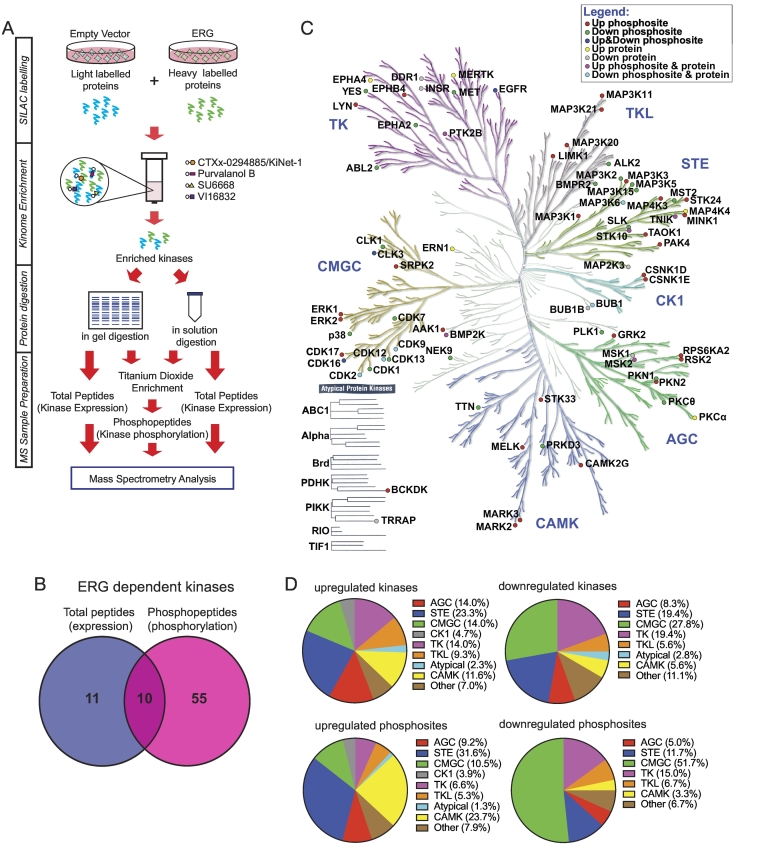
Figure 1.
Profiling the ERG-regulated prostate cancer kinome. (A) Workflow for quantitative MS-based kinomic profiling. (B) Summary of kinome profiling data. Number of kinases identified to exhibit a ≥1.2-fold change in expression, phosphorylation or both in ERG overexpressing cells, when compared to the empty vector control, in both biological replicates. (C) Distribution of ERG-regulated kinases over the human kinome. Kinome tree generated using Kinome Render [34]. Illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com). (D) Proportional representation of ERG-regulated kinases. Kinases up- and/or down-regulated by ERG in terms of expression and/or phosphorylation from (C) were categorized into their kinase families (top). Proportional representation of regulated kinase phosphosites. Each ERG-regulated phosphosite was categorized into the family of its respective kinase (bottom).