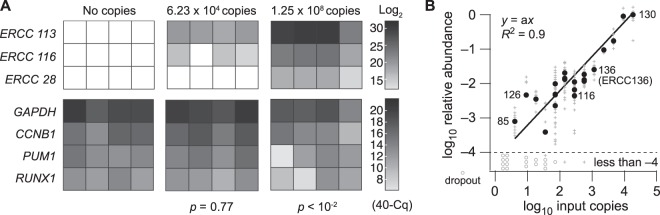
Figure 4.
Optimized ERCC spike-in dilutions assess poly(A) PCR sensitivity and dynamic range without suppressing cDNA amplification of endogenous transcripts. (A) 100 pg RNA was supplemented with ERCC Mix 1 at the indicated dilutions and amplified via optimized poly(A) PCR. ERCC and endogenous gene abundances were measured by qPCR, and data are shown in grayscale as the inverse quantification cycle (40–Cq) from n = 4 amplification replicates. Negative effects of the ERCC spike-ins on endogenous genes (lower) were assess by two-way ANOVA with replication. (B) ERCC Mix 1 (6.23 × 104 copies) was spiked into 100 pg RNA and amplified via optimized poly (A) PCR. Proportional abundance of ERCC standards was estimated with a seven-log dilution series from purified qPCR end products. Data are shown as the median 40–Cq (black) for 22 ERCC spike-in standards from n = 8 amplification replicates (gray) with undetected “dropouts” reported below (circles).