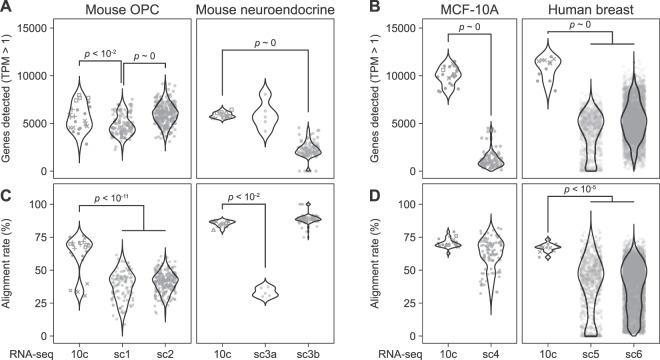
Figure 8.
Increased gene detection and improved exonic alignment rates for 10cRNA-seq compared to scRNA-seq. (A) Detection of murine Ensembl genes for mouse oligodendrocyte precursor cells (OPCs) and lung neuroendocrine-derived cells. (B) Detection of human Ensembl genes for MCF-10A cells and human breast cells. (C) Exonic alignment rate comparison for OPCs and lung neuroendocrine-derived cells. (D) Exonic alignment rate comparison for MCF-10A cells and human breast cells. Public scRNA-seq data were obtained from the indicated accession numbers: sc1 = GSE75330, sc2 = GSE60361, sc3a = GSE103354 (plate-based), sc3b = GSE103354 (droplet-based), sc4 = GSE66357, sc5 = GSE113197, sc6 = PRJNA396019. 10cRNA-seq data were aggregated from independent 10-cell samples (circles) and 10-cell equivalents from pool-and-split controls. Pool-and-split controls from the same day are indicated with non-circular markers corresponding to the shared day. Pairwise differences between 10-cell and single-cell methods were assessed by permutation test.