Abstract
Low-passage, serum-free cell lines cultured from patient tumour tissue are the gold-standard for preclinical studies and cellular investigations of glioblastoma (GBM) biology, yet entrenched, poorly-representative cell line models are still widely used, compromising the significance of much GBM research. We submit that greater adoption of these critical resources will be promoted by the provision of a suitably-sized, meaningfully-described reference collection along with appropriate tools for working with them. Consequently, we present a curated panel of 12 readily-usable, genetically-diverse, tumourigenic, patient-derived, low-passage, serum-free cell lines representing the spectrum of molecular subtypes of IDH-wildtype GBM along with their detailed phenotypic characterisation plus a bespoke set of lentiviral plasmids for bioluminescent/fluorescent labelling, gene expression and CRISPR/Cas9-mediated gene inactivation. The cell lines and all accompanying data are readily-accessible via a single website, Q-Cell (qimrberghofer.edu.au/q-cell/) and all plasmids are available from Addgene. These resources should prove valuable to investigators seeking readily-usable, well-characterised, clinically-relevant, gold-standard models of GBM.
Subject terms: Cancer models, Cancer stem cells, CNS cancer, Cancer stem cells
Introduction
Glioblastoma (GBM; WHO grade IV astrocytoma) is the most common, and most lethal, primary malignant adult brain cancer1. Despite surgery, radiotherapy and temozolomide chemotherapy, GBM patients have a median survival of <15 months and a 5-year survival rate of only 10%2. GBM almost always recurs following treatment and is then rapidly fatal with no current mainstay therapy effectively altering the course of the disease1. Two factors that contribute to treatment failure are the heterogeneity of GBM – in part represented by tumour cells with distinctly different patterns of gene expression3,4 (proneural, classical and mesenchymal GBM) – and the presence of glioma stem cells (GSCs), chemo-radiotherapy resistant cells that are able to reconstitute tumour architecture and are believed to be responsible for disease progression and recurrence5–7. Preclinical cell line and animal models that recapitulate these features accurately are essential if our understanding of GBM biology is to improve and more effective strategies to treat this almost uniformly fatal disease are to be developed.
Low-passage, serum-free, patient-derived GBM cell lines have proven to be the gold-standard in this regard. Methods to initiate and propagate such lines from patient tumour tissue are well established8–10. These approaches not only capture the genetic diversity of human GBM, but also help preserve the genomic fidelity of such cells and enrich for the presence of GSCs, enhancing their tumour-initiating capacity11. Significantly, xenograft tumours that arise from these cells are invasive and exhibit the histological hallmarks of high grade glioma, including hypercellularity, nuclear atypia and the presence of mitotic figures, with or without microvascular proliferation and (palisading) necrosis12. In essence, such cell lines are a highly relevant model for studying GBM, capturing the heterogeneity of this disease and recapitulating its cancer stem cell biology.
Despite their greater relevance for understanding human GBM, more widespread use of low-passage, serum-free, patient-derived cell lines in much GBM research appears not to be restricted by technical challenges but rather issues associated with the acquisition of patient tissue, unfamiliarity with existing lines, and/or much greater familiarity with alternative long-established (although poorly-representative) cell line models of GBM. Here we aspire to increase the wider use of low-passage, serum-free, patient-derived GBM cell lines amongst the cancer and cell biology research community by presenting a reference set - a curated, practical-size collection of genetically-diverse GBM cell lines established from different molecular subtypes of GBM - that form invasive xenograft tumours in immunodeficient mice. All lines have been characterised in significant practical detail, for interrogation and immediate use by researchers, with all data accessible via a user-friendly website. The availability of this data for in silico analysis should facilitate selection of lines suitable for hypothesis testing while the size, genetic diversity and subtype representation of the collection provides incentive for acquisition of the entire set to broadly and meaningfully inform studies of GBM.
In addition, we provide a set of tools to facilitate functional analysis of the lines. Lentiviral plasmids for bioluminescent and/or fluorescent labelling, expressing genes of interest and CRISPR/Cas9-targeted gene inactivation are presented. These further enhance the utility of the collection for investigating the biology of GBM.
Results
Establishment of a diverse set of low-passage, serum-free GBM cell lines from patient tumour tissue
Between November 2009 and June 2011, we established GBM cell lines as serum-free, adherent cultures from tissue collected from patients undergoing surgery at the Royal Brisbane and Women’s Hospital with a success rate of 75%. Twelve morphologically-diverse, rapidly-growing cell lines, that formed xenograft tumours when injected intracranially into NOD/SCID mice, were selected as a GBM reference collection for subsequent detailed characterisation. The patient cohort from which the cell lines were established had a median age of 70 years (range 48–84 years) and included 7 males and 5 females (Table 1). Eleven had primary GBM and one had recurrent GBM. Seven were right-sided tumours (frontal, parietal, temporal or occipital), five were left-sided (frontal or parietal). De-identified pathology reports for each tumour are attached as Supplementary Data S1. Overall survival ranged from 36 days to 7 years from the date of diagnosis of GBM – median survival was 5 months (Supplementary Fig. S1).
Table 1.
Patient demographics.
| Patient | Age (years) | Gender | Tumour type | Tumour site | Survival (days) |
|---|---|---|---|---|---|
| BAH1 | 75 | Female | Primary GBM | Right frontal | 94 |
| FPW1 | 68 | Male | Primary GBM | Right temporal | 242 |
| HW1 | 54 | Female | Primary GBM | Right frontal parietal | 89 |
| JK2 | 75 | Male | Primary GBM | Right frontal | 178 |
| MMK1 | 80 | Female | Primary GBM | Right temporal | 334 |
| MN1 | 84 | Female | Primary GBM | Left frontal | 36 |
| PB1 | 57 | Male | Primary GBM | Left frontal | 39 |
| RKI1 | 57 | Female | Primary GBM | Left temporal | Alive (7 years) |
| RN1 | 56 | Male | Primary GBM | Left temporal | 243 |
| SB2b | 48 | Male | Recurrent GBM | Right parietal | 420 |
| SJH1 | 72 | Male | Primary GBM | Left temporal | 45 |
| WK1 | 77 | Male | Primary GBM | Right parietal occipital | 121 |
Abbreviations: GBM, glioblastoma.
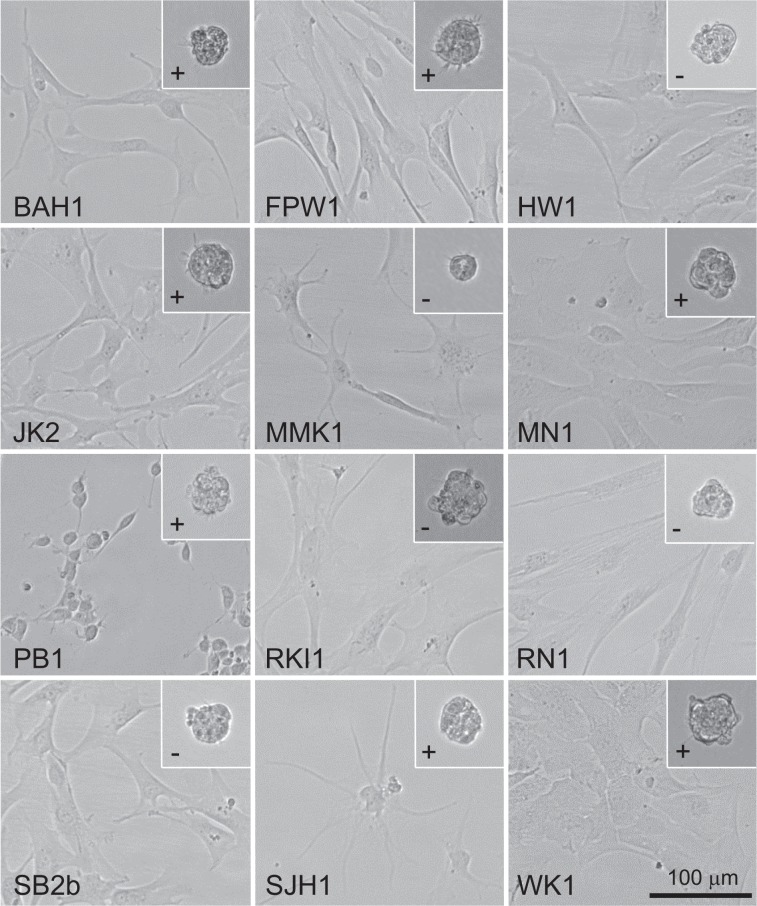
Low-passage, serum-free, patient-derived cell lines were established either from discrete pieces of tumour tissue or pooled tissue collected by an ultrasonic surgical aspirator13. Tumour tissue from both sources was mechanically and enzymatically dissociated and cultured on matrigel-coated plastic in serum-free RHB-A medium supplemented with epidermal growth factor and fibroblast growth factor 2. Medium was changed every three to four days and cells were washed with phosphate-buffered saline to remove tissue debris. Cells were passaged into larger flasks when nearing confluence and the first freezings were made from 75 cm2 flasks after continuous culture ranging from 14 to 30 days. The resultant cell lines exhibited a variety of cell morphologies (Fig. 1 and Supplementary Fig. S1). This heterogeneity was most marked among the different cell lines although also evident within many of them. Doubling times at passage 10 ranged between 38 and 109 hours (Table 2). Seven of the lines grew readily as tumourspheres when cultured without matrigel (Fig. 1); the other five lines formed tumourspheres poorly or not at all. This behaviour did not seem to correlate with GBM subtype.
Figure 1.
A diverse set of low-passage, serum-free, patient-derived GBM cell lines exhibiting a range of cell morphologies. Bright field images of GBM cell lines growing as adherent cultures on matrigel (main images) or as non-adherent tumourspheres (inset). Plus signs (+) indicate cell lines that readily form tumourspheres; minus signs (−) indicate cell lines that form tumourspheres poorly or not at all. Scale bar, 100 μm.
Table 2.
GBM cell line phenotypes.
| Patient | Reference number | Doubling time (hours) | IDH1 status | MGMT methylation | Xenograft median survival (days) |
|---|---|---|---|---|---|
| BAH1 | QIMR-B001 | 79.5 +/− 3.3 | WT | Methylated | 210 +/− 8 |
| FPW1 | QIMR-B002 | 48.1 +/− 4.7 | WT | Unmethylated | 196 +/− 4 |
| HW1 | QIMR-B003 | 55.8 +/− 2.7 | WT | Methylated | 174 +/− 14 |
| JK2 | QIMR-B004 | 94.2 +/− 6.0 | WT | Unmethylated | 147 +/− 9 |
| MMK1 | QIMR-B005 | 52.4 +/− 4.5 | WT | Unmethylated | 157 +/− 15 |
| MN1 | QIMR-B006 | 44.9 +/− 1.0 | WT | Unmethylated | 258 +/− 20 |
| PB1 | QIMR-B007 | 79.4 +/− 6.3 | WT | Unmethylated | 71 +/− 1 |
| RKI1 | QIMR-B008 | 72.9 +/− 5.3 | WT | Methylated* | 248 +/− 6 |
| RN1 | QIMR-B009 | 37.5 +/− 1.9 | WT | Unmethylated | 81 +/− 2 |
| SB2b | QIMR-B010 | 108.7 +/− 6.9 | WT | Methylated | 120 +/− 4 |
| SJH1 | QIMR-B011 | 67.3 +/− 4.7 | WT | Unmethylated | 148 +/− 2 |
| WK1 | QIMR-B012 | 46.2 +/− 1.0 | WT | Unmethylated | 150 +/− 2 |
Abbreviations: *homozygous SNP rs16906252; IDH1, isocitrate dehydrogenase 1; MGMT, methyl guanine methyltransferase; WT, wild type.
Expression of O-6-methylguanine-DNA methyltransferase (MGMT) from the MGMT gene confers resistance in GBM cells to the alkylating chemotherapy agent temozolomide. Methylation of the MGMT promoter silences MGMT transcription. The MGMT promoter was found to be methylated in four of the GBM cell lines, one of which contained a homozygous single nucleotide polymorphism (rs16906252) (Table 2).
Mutation of the gene encoding isocitrate dehydrogenase (NADP(+)) 1, (IDH1) at arginine 132 or IDH2 at arginine 172 is associated with secondary GBM, GBM that has arisen from lower grade glioma. All lines were found to be wild type for IDH1and IDH2 so can be regarded as glioblastoma, IDH-wildtype.
Each cell line was short tandem repeat profiled (Supplementary Table S1). All STR profiles were unique and alleles for the AMEL locus matched the gender of the patients from which each cell line was established. There was no evidence of significant relatedness with known cell lines or evidence of cross-contamination among the 12 lines. All lines tested mycoplasma-free.
To complement the collection of GBM cell lines, we created a set of lentiviral plasmids for ectopic gene expression or CRISPR/Cas9-mediated gene inactivation (Supplementary Table S2). Expression plasmids contain the chimaeric human CMV enhancer/chicken beta-actin promoter for strong gene expression in GBM cells and confer resistance to puromycin, hygromycin B or G418. By default, they contain the firefly luciferase gene to generate bioluminescent cell lines. Lentiviral CRISPR/Cas9 plasmids were derived from lentiCRISPRv2 (Supplementary Fig. S2) and confer resistance to puromycin, hygromycin B, G418 or blasticidin S. To aid the establishment of transformed cell lines with these vectors we determined the sensitivity of each of the GBM cell lines to these antibiotics (Table 3).
Table 3.
GBM cell line antibiotic sensitivity (μg/ml).
| GBM cell line | Puromycin | Hygromycin B | G418 | Blasticidin S |
|---|---|---|---|---|
| BAH1 | 0.5 (5) | 100 | 200 | 1 |
| FPW1 | 0.5 | 200 (9) | 300 (9) | 4 |
| HW1 | 0.4 | 200 | 300 | 2 |
| JK2 | 0.4 | 100 | 300 | 2 |
| MMK1 | 0.5 | 100 | 200 | 2 |
| MN1 | 0.4 (5) | 200 | 200 | 2 |
| PB1 | 0.2 (5) | 200 | 200 | 1 |
| RKI1 | 0.5 | 200 (10) | 200 (10) | 4 |
| RN1 | 0.5 | 100 | 200 | 1 |
| SB2b | 0.5 (5) | 200 | 400 | 1 |
| SJH1 | 0.5 | 200 | 300 | 1 |
| WK1 | 0.5 | 200 (9) | 400 | 4 |
Notes: 7 days unless specified ().
Characterisation of GBM cell line intracranial tumourigenicity in NOD/SCID mice
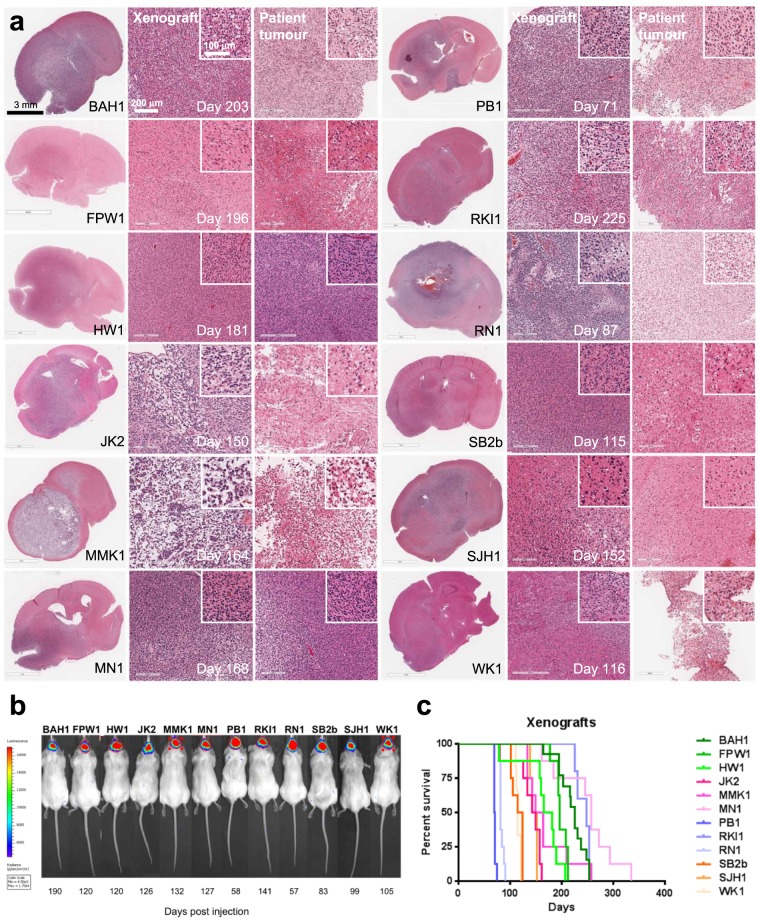
To investigate the tumourigenicity of each cell line, 1.5 × 105 cells were injected intracranially into the right striatum of cohorts of 5-week old NOD/SCID mice. All cell lines formed invasive tumours with similar histology to the patient tumours from which they were established (Fig. 2a), including evidence of focal necrosis and vascular proliferation. No clear correlation though was apparent between xenograft histology and median survival time.
Figure 2.
Bioluminescent xenograft tumour formation in NOD/SCID mice. (a) Comparative histology of xenograft tumours formed by the GBM cell lines and the patient tumours they were established from. First column, haematoxylin and eosin (H&E)-stained, coronal sections of mouse brains showing invasive xenograft tumours. Scale bar, 3 mm. Comparative H&E-stained sections of matched xenograft (middle column) and patient tumours (right column). Scale bar 200 μm, inset 100 μm. (b) Bioluminescent intracranial xenograft tumours produced by the GBM cell lines expressing firefly luciferase. Stably-transduced lines were created by transduction with lentivirus containing pF CAG luc puro and selection with puromycin. Time to develop the bioluminescent xenograft tumours detected is shown below each mouse. (c) Kaplan-Meier survival curves for NOD/SCID mice harbouring xenografts initiated by intracranial injection of each of the GBM cell lines.
To enable tumour growth to be monitored by bioluminescence, each cell line was also transduced with a lentiviral transfer plasmid expressing luciferase from a chimaeric CMV enhancer/chicken beta actin promoter. Luciferase-expressing cells generated bioluminescent xenograft tumours with tumour-bearing mice having median survival times ranging from 2.5–9 months (Table 2 and Fig. 2b,c). Luciferase expression was not observed to alter xenograft median survival.
Genomic characterisation of the GBM cell lines
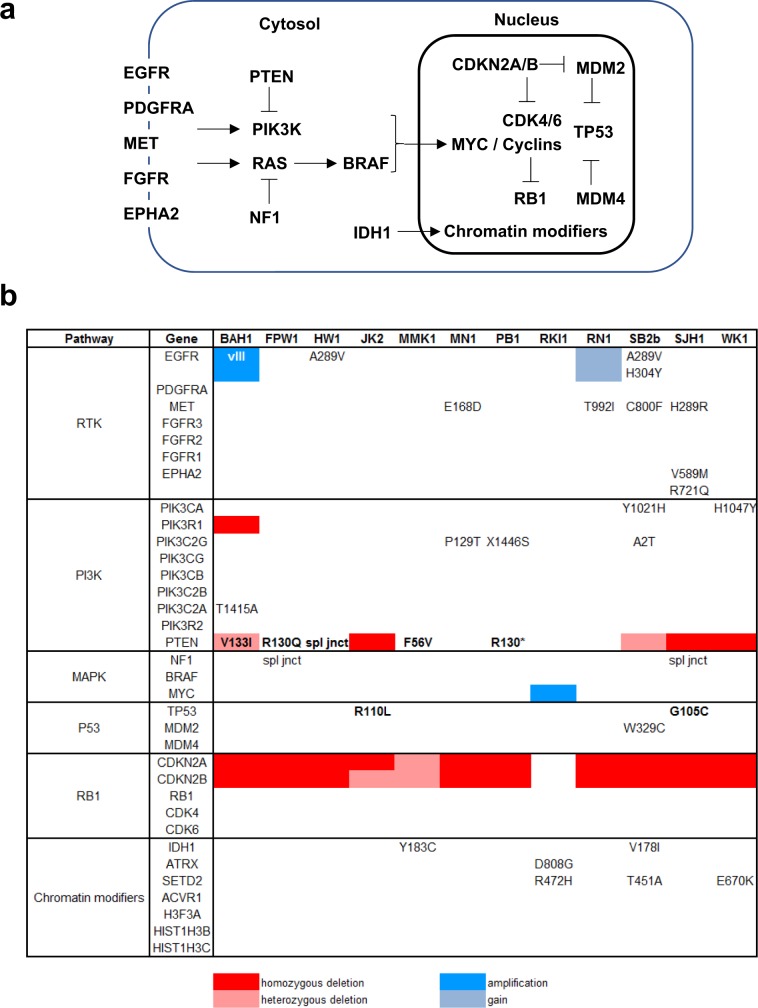
To catalogue the genotype of each GBM cell line, we performed exome sequencing. Genomic alterations characteristic of GBM were common among the 12 lines (Fig. 3a,b). Most frequent was homozygous deletion of the tumour suppressor CDKN2A, exhibited by 10 (83%) of the lines. Homozygous deletion of PTEN was detected in three lines and homozygous single nucleotide variants (SNVs) that were predicted to be pathogenic14 were present in five others. TP53 nonsynonymous SNVs were infrequent, with a homozygous predicted pathogenic missense SNV (G105C) in SJH1 and a homozygous neutral missense SNV (R110L) in JK2. At least half of the lines contained potential pathogenic changes in GBM-associated receptor tyrosine kinase (RTK) genes. Amplification of the oncogenic vIII variant of EGFR (EGFRvIII) was present in one of the lines (BAH1) and copy number gain of wild type EGFR was detected in another (RN1). Two further lines (HW1 and SB2b) contained pathogenic SNVs in EGFR that have only been reported previously in GBM15. Missense SNVs also were detected in MET and EPHA2, including a substitution in the latter that creates a variant (EphA2 R721Q) with altered signalling. Amplification of MYC, detected in only 1.5% of GBM16,17, was present in the cell line established from a long-term survivor (RKI1). Nonsynonymous single nucleotide variant calls for each of the cell lines, their zygosity and predicted measures of their pathogenicity are listed as Supplementary Data S2.
Figure 3.
Genomic profiling of the GBM cell lines. (a) Pathways most frequently affected by genomic changes in GBM showing the relationships of the genes involved. (b) Single nucleotide variants (SNVs), intragenic deletions and gene copy number changes in genes in these pathways detected in the GBM cell lines. SNVs shown in bold are homozygous, otherwise heterozygous. Blanks indicate wild type genes.
Gene expression and molecular subtype analysis of matched patient tumours, cell lines and xenografts
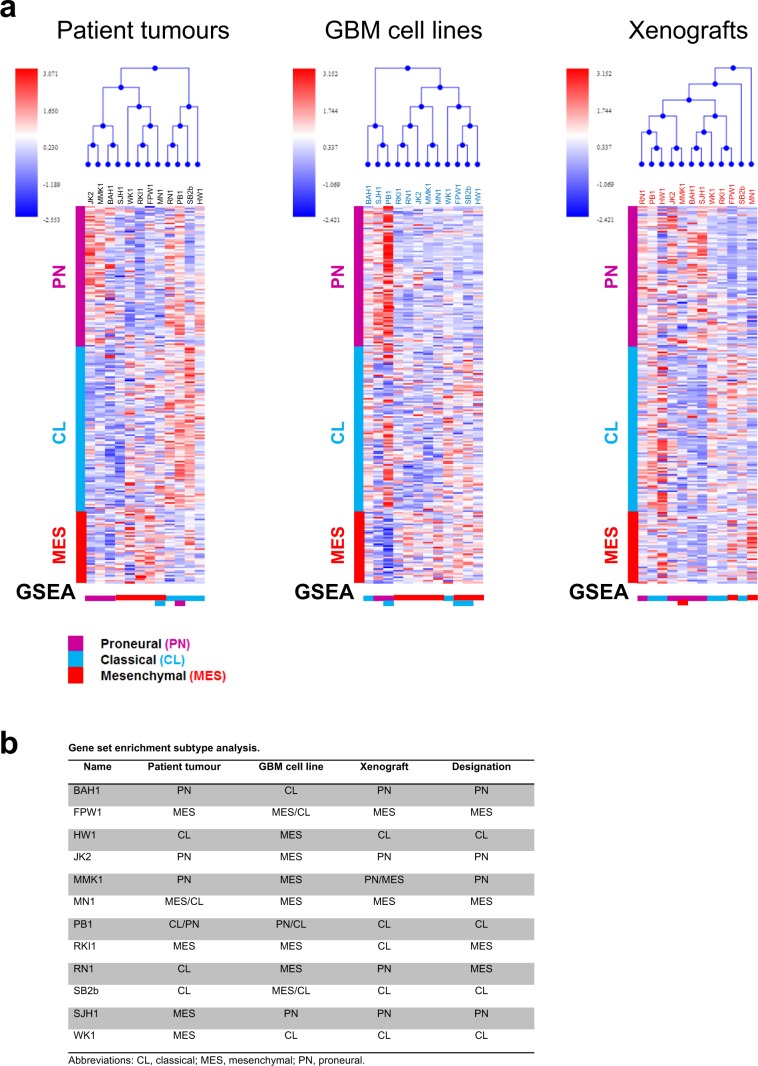
Although GBM is a genetically and phenotypically heterogeneous cancer, distinct molecular subtypes can be distinguished, including on the basis of gene expression. The most widely adopted molecular classification system uses an 840-gene signature to distinguish proneural, neural, classical and mesenchymal GBM4. These subtypes resemble distinct normal neural cell types, have different prognoses, respond differently to treatment and are associated with characteristic genetic alterations4, thus these distinctions appear to have patho-biological significance. Since the introduction of this classification schema by The Cancer Genome Atlas consortium, reservations have been expressed concerning the identity of the neural GBM subtype. Greatest support in the literature is found for the proneural, classical and mesenchymal subtypes18, with the suggestion that neural GBM likely represents normal brain tissue containing a small percentage of tumour cells19, an observation that could account for this subtype lacking characteristic genetic alterations20. To molecularly subtype the tumour tissue from which our GBM cell lines were derived, we measured RNA expression and performed hierarchical cluster analysis (Fig. 4a) and gene set enrichment analysis (GSEA) (Supplementary Fig. S3) using the classifier gene sets for proneural, classical and mesenchymal GBM. This showed our GBM cell lines were established from tumours of each of these molecular subtypes of GBM. Consistent with the finding that individual tumours may contain subpopulations of tumour cells of different molecular subtypes21,22, and reflecting the heterogeneity of this disease, several of the tumour specimens exhibited gene expression profiles of more than one GBM subtype (Supplementary Fig. S3). Inclusion of the neural signature in the GSEA analysis showed several of the specimens (BAH1, HW1, RKI1 and SJH1) also exhibited significant expression of neural genes, consistent with the sample containing both tumour and normal tissue and this was confirmed by histological examination of the tumour tissue adjacent to that used to extract RNA (data not shown).
Figure 4.
Molecular subtyping of matched patient tumours, GBM cell lines and xenografts. (a) Hierarchical cluster analysis (top), corresponding heatmaps (middle) and gene set enrichment analysis (GSEA) (bottom) of patient tumours, derived GBM cell lines and matched murine xenografts initiated by the cell lines identifying gene expression patterns characteristic of proneural, classical and mesenchymal GBM. (b) Summary of GSEA results.
To investigate whether the GBM cell lines retained the phenotype of the tumour from which they were derived, we similarly analysed both cultured cells and the xenografts they produced (Fig. 4a and Supplementary Fig. S3). We found the majority of GBM cell lines (7/12) exhibited a mesenchymal gene expression signature. When transplanted into NOD/SCID mice, however, most cell lines (7/12) produced xenografts of the same molecular subtype as the tumour from which they arose, including five that exhibited a different molecular subtype when grown in culture. Cell lines derived from mesenchymal patient tumours most often (4/5) gave rise to xenografts of a different subtype (most often classical GBM) despite retaining their mesenchymal subtype in culture. Notably, in only two cases was the same molecular subtype observed for patient tumour, GBM cell line and xenograft: MN1 (mesenchymal) and SB2b (classical).
Our final designation of the subtype of each GBM cell line and a summary of the results for patient tumours, cell lines and xenografts is shown in Fig. 4b. Most designations match the subtype of the primary tumour. In the three that did not (RN1, SJH1 and WK1), the significantly higher gene set enrichment scores for the cell lines and/or xenografts led us to designate these as mesenchymal, proneural and classical respectively.
We also noted MGMT expression in the GBM cell lines was consistent with MGMT promoter methylation status. Interestingly, although the MGMT promoter was unmethylated in the MN1 cell line, no MGMT expression was detected. As exome sequencing showed there were no alterations in the MGMT gene, this indicated factors besides promoter methylation also determine MGMT expression.
As a reference resource, microarray gene expression data for all patient tumours, GBM cell lines and xenografts are provided as Supplementary Data S3. RNAseq analysis of gene expression for the GBM cell lines also is provided as Supplementary Data S4. Microarray raw data have been deposited with the GEO repository with the series accession number GSE118793 and RNAseq raw data is available for download from the NCBI sequence read archive with SRA accession number PRJNA508446.
GBM-associated protein expression
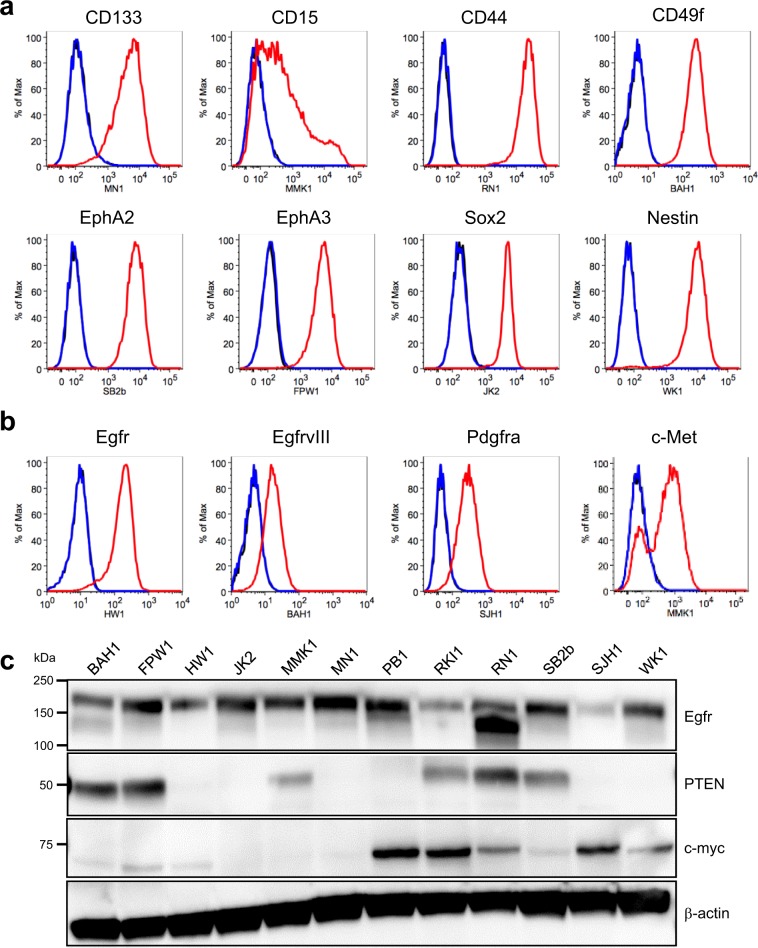
Several cellular proteins are used as markers of glioblastoma stem and progenitor cells (GSCs) and are employed to isolate these cells by flow cytometry or affinity capture. We therefore investigated the expression of GSC markers by flow cytometry in cultured GBM cell lines (Fig. 5a and Supplementary Data S5), including the GSC marker CD133, whose protein level is independent of CD133 mRNA expression23, CD15, CD44, CD49f, EphA224 and EphA325, and the progenitor cell markers Sox2 and Nestin7. All were detected among the 12 GBM cell lines, consistent with the culture conditions maintaining the presence of GSCs. Some markers were detected only in some cell lines, others in a subset of cells in a given line. We also investigated expression of several other GBM-associated RTKs, in addition to EphA2 and EphA3, including Egfr, EgfrvIII, Pdgfrα and c-Met (Fig. 5b and Supplementary Data S5). Our findings were in accord with our exome sequencing (EgfrvIII) and mRNA expression analysis, with widespread expression of Egfr and EphA2 and more restricted expression of the other RTKs. Widespread expression of Egfr also was detected by western blot (Fig. 5c and Supplementary Fig. S4); interestingly EGFR copy number gain in the RN1 cell line was associated with strong expression of a low molecular weight form of Egfr which did not appear to be EgfrvIII as this was not detected by flow cytometry using an EgfrvIII-specific monoclonal antibody (Supplementary Data S5). Similarly, western blot (Fig. 5c and Supplementary Fig. S4) confirmed lack of expression of PTEN in lines in which PTEN was homozygously deleted (JK2, SJH1 and WK1), contained a splice junction mutation (HW1) and showed gain of a premature STOP codon (PB1, R130*). PTEN expression also was not detected in a line (MN1) in which no mutations were detected in the coding sequence or splice junctions of the PTEN gene by exome sequencing. Increased expression of c-myc in the cell line (RKI1) with MYC amplification also was confirmed by western blot (Fig. 5c and Supplementary Fig. S4). Interestingly, several other lines (PB1, SJH1, RN1 and WK1) also showed evidence of increased c-myc expression, suggesting they may be c-myc-driven GBMs. Protein expression (or lack thereof) from other GBM-associated genes, including CDKN2A, CDKN2B, FGFR1, NF1, TP53, RB1 and IDH1 (Fig. 3b), was confirmed by proteomic analysis (Supplementary Data S6).
Figure 5.
GBM-associated protein expression in the GBM cell lines. Representative flow cytometry analysis of (a) stem cell markers and (b) selected receptor tyrosine kinases in the GBM cell lines cultured as adherent monolayers on matrigel. (c) Western blot analysis of Egfr, Pten and c-myc expression in the GBM cell lines. kDa, kilodalton.
Discussion
Many preclinical and laboratory investigations of human GBM use cell lines. Historically these have been maintained in serum-supplemented medium that, while providing growth factors and other mitogens, introduces genotoxic stress. This, coupled with extensive passaging, produces cell line and xenograft models whose phenotypes diverge significantly from their tumour of origin. Low passage, patient-derived GBM cell lines that are maintained in serum-free medium supplemented with EGF and FGF2, have more recently been adopted by many investigators to circumvent these pressures and produce models that more accurately recapitulate the cardinal features of GBM11. Despite their clear benefits and ease of use, the wider adoption of such cell lines is often restrained by several factors. Initiating the necessary collaborations, applying for human research ethics approval and establishing cell lines from patient tumour tissue requires the investment of significant time and effort. Alternatively, sourcing such lines from other laboratories often comes with limited understanding regarding the origin and phenotype of the cells received as well as a lack of familiarity with working with them. Having experienced both scenarios, we have tried to increase investigator access to, and confidence with using, such cell lines by compiling a meaningfully-sized reference collection of extensively-characterised, low-passage, serum-free, patient-derived GBM cell lines, along with bespoke tools for working with them, which we hope will lead to more widespread use of this more relevant resource for investigating human GBM. While a number of laboratories have reported GBM cell line and mouse xenograft models of GBM, and a larger publicly-available, partially-phenotyped, GBM cell line collection exists26, we believe the practical size of our collection, coupled with the extent and practical-focus of its genotypic and phenotypic characterisation, meets a need in the GBM research community for a reference collection suitable for acquiring en bloc for laboratories yet to make the switch to using such cell lines or alternatively as a reliable source of individual lines with defined phenotypes and genotypes from which other laboratories can supplement their existing resources.
During the preparation of this manuscript, a revised TCGA classification system was reported for IDH-wild type GBM27. This retains the proneural, classical and mesenchymal nomenclature (and drops the neural subtype) and uses a smaller number of genes (50 for each subtype) to subtype GBM. When this smaller classifying gene set was used to subtype our tumour samples (all IDH-wild type), all were classified with much lower (and often nonsignificant) normalised enrichment scores. In light of this experience, there may be advantages in using the original TCGA classification system, with its larger number of classifier genes, to distinguish proneural, classical and mesenchymal GBM, with the inclusion of the neural classifier to identify samples that contain few tumour cells.
The key finding of our subtype analysis of matched patient tumours, GBM cells lines and xenografts was that low-passage, serum-free cell lines most often gave rise to xenografts of the same molecular subtype as the patient tumours from which they were derived, although frequently exhibited a different GBM subtype during in vitro culture. That most of our GBM cell lines exhibited a mesenchymal gene expression signature, regardless of the subtype of their tumour of origin, was striking yet is an inconsistent finding in the literature4,26,28. That serum-free cell lines derived from mesenchymal patient tumours often produced xenografts of a different subtype, despite retaining their mesenchymal subtype in culture, has been observed independently26,28. Yet to be determined is whether the apparent subtype switching is the result of the emergence of pre-existing subpopulations of cells29 or represents inherent plasticity28. While our patient tumours may contain GBM cells of more than one molecular subtype, which could account for the subtype “switch” observed between patient tumours and the derived GBM cell lines (in vitro culture potentially selecting for one subtype over another), subtype differences between the cell lines and the xenografts they produce suggest some GBM cells may be plastic and that their phenotype can change in response to their environment. We anticipate that the heterogeneity inherent in this collection of low-passage, serum-free GBM cell lines will be helpful for resolving this and other issues of GBM cell biology.
Interestingly, only one EgfrvIII-expressing and one Egfr-amplified cell line was established despite EGFR alterations occurring in over half of GBM tumours20. This is a common observation and may be related to a requirement in vivo that is lost in culture. As we observed that omission of recombinant Egf from the culture medium increased expression of EgfrvIII in the BAH1 cell line (data not shown), it may be worthwhile to routinely establish GBM cell lines both in the presence and absence of Egf in the culture medium in order to attempt to increase the frequency at which Egfr/EgfrvIII-amplified cell lines are established.
Investigations to characterise our GBM cell lines were performed between passage 8 and 15, at which time we considered them “established”. Each line has been passaged at least 30 to 50 times now without notable change in tumourigenicity, growth rate, morphology or antibiotic sensitivity. While proneural tumours are typically less aggressive, our proneural xenograft-forming cell lines did not have the longest median xenograft survival (166 +/− 9 days for proneural xenografts versus 162 +/− 8 days for classical and mesenchymal combined) suggesting this behaviour may not apply to xenograft models. However, our sample size is small and larger numbers will be required to confirm this.
In our hands, as well as in other laboratories, the GBM cell lines have proven useful for a variety of purposes. These include the identification and validation of GBM- and GSC-associated therapeutic targets25,30–32, investigations of GBM metabolism33, microRNA function in GBM34, GBM sensitivity to ionising radiation35, PET/MRI imaging of GBM36,37, GBM cell response to environmental stiffness38 and the evaluation of novel agents for treating GBM39–45. In light of this demonstrated utility, we anticipate this GBM cell line reference collection, and accompanying set of lentiviral plasmids, will both prove to be valuable resources for the scientific research community.
Methods
Tumour tissue and cell culture
Patient tumour tissue was collected from patients undergoing surgery at the Royal Brisbane and Women’s Hospital (RBWH), with informed consent and human ethics approval from the QIMR Berghofer Medical Research Institute and RBWH human research ethics committees. All human studies have been performed in accordance with the ethical standards laid down in the 2013 version of the 1964 Declaration of Helsinki. Tumour tissue was examined by a neuropathologist (TR) to determine tumour type and grade. Patient-derived cell lines were established as reported previously13. These were cultured as adherent monolayers in matrigel (BD Biosciences)-coated vessels using RHB-A stem cell culture medium (StemCells Inc) supplemented with 20 ng/ml EGF (Gibco) and 10 ng/ml FGFb (Gibco) and containing 100 U/ml penicillin and 100 μg/ml streptomycin, or as tumourspheres using StemPro NSC SFM (Invitrogen). Cells were cultured in 5% CO2/95% humidified air at 37 °C. All cell lines were routinely tested for mycoplasma and confirmed to be mycoplasma-free.
Short tandem repeat profiling
Short tandem repeat (STR) profiling to establish the unique genomic identity of each cell line and exclude cross-contamination was performed by the QIMR Berghofer Medical Research Institute Analytical Facility. Alleles for 10 human loci (THO1, D21S11, D5S818, D13S317, D7S820, D16S539, CSFIPO, AMEL, vWA and TPOX) were assayed and compared with the ATCC STR database (https://www.atcc.org/STR_Database.aspx) using the standard match threshold of 80%.
MGMT promoter methylation detection
MGMT promoter methylation status was determined by bisulphite pyrosequencing as described46.
IDH1 mutation status
The mutation status of Idh1 at R132 was determined by PCR amplification of IDH1 from genomic DNA using the oligonucleotide primers IDH1 forward: CGGTCTTCAGAGAAGCCATT and IDH1 reverse: GCAAAATCACATTATTGCCAAC and direct Sanger sequencing of the resultant PCR product using the IDH1 forward primer. IDH1 and IDH2 mutation status also was confirmed by exome sequencing.
Proliferation assays and doubling time
Cells were seeded at a density of 2 × 103 cells per well in 100 μl of medium in matrigel-coated 96-well plates and MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] proliferation assays were performed using a CellTiter 96 AQueous One Solution cell proliferation assay kit (Promega). Cells were incubated with 20 μl of MTS reagent and incubated for one hour at 37 °C. The ratio of absorbance in the test wells to control wells (containing culture medium and no cells) was calculated as relative MTS activity and used as a surrogate for the number of viable cells in culture. Average relative MTS activity from three replicate wells was plotted against time and the doubling time calculated using GraphPad Prism software v.6.0 (GraphPad software, LaJolla, CA, USA).
Plasmid construction
Lentiviral expression plasmids were derived from pF_U_MCS_SV40Puro and pF_U_G147EV16_PGK_Hygro47 (a gift from John Silke). The chimaeric human CMV enhancer/chicken beta-actin promoter from pCAGIG48 (a gift from Connie Cepko) was subcloned in place of the ubiquitin promoter in both vectors and firefly luciferase was subcloned from pGL2 Basic (Promega) to generate pF CAG luc puro and in place of G147EV16 to produce pF CAG luc hygro. pF CAG luc IRES neo was created by cloning an IRES-neo cassette into pF CAG luc hygro in place of PGK-hygro. The IRES sequence was amplified by PCR from pIRES2-DsRed-Express (Clontech) and the neo sequence was amplified from pSuperior neo + GFP (OligoEngine). Genes of interest can be subcloned between the unique BamHI and NheI restriction sites in place of luciferase. pF CAG BFP2 IRES neo was created by cloning PCR-amplified BFP2 from pBAD-mTagBFP249 (a gift from Vladislav Verkhusha, Addgene plasmid #34632) between the BamHI and NheI restriction sites of pF CAG luc IRES neo.
CRISPR/Cas9 plasmids were constructed from lentiCRISPRv250 (a gift from Feng Zhang, Addgene plasmid #52961) and are designed to confer resistance to puromycin, hygromycin B, G418 or blasticidin S. These were created by first destroying the MluI restriction site of lentiCRISPRv2 at nucleotide position 231 by MluI digestion followed by Klenow end-filling and re-ligation to create lentiCRISPRv2 puro, then cloning P2A-antibiotic resistance cassettes between the MluI and BamHI restriction sites of lentiCRISPRv2 puro. The P2A sequence was from lentiCRISPRv2, the hygromycin resistance gene cassette from pF CAG luc hygro, the neomycin resistance gene cassette from pF CAG luc IRES neo, and the blasticidin resistance gene cassette from pcDNA6TR (Invitrogen). Gene fusions were created by overlapping PCR. Short guide sequences can be cloned between the BsmBI restriction sites as previously described50. Cas9-only and sgRNA-only derivatives were constructed as shown in Supplementary Fig. S2. All plasmids have been deposited with, and their sequence verified by, Addgene (see Supplementary Table S2 for Addgene reference numbers or addgene.org/Brett_Stringer/). Instructions for using the lentiCRISPRv2 plasmids are as described by the Zhang laboratory (https://media.addgene.org/cms/filer_public/53/09/53091cde-b1ee-47ee-97cf-9b3b05d290f2/lenticrisprv2-and-lentiguide-oligo-cloning-protocol.pdf).
Lentivirus production
Lentivirus was produced in 10 cm diameter petri dishes from 70–80% confluent HEK293FT cells transfected with 3 μg lentiviral plasmid, 2 μg pVSV-G (Addgene #8454) and 5 μg psPAX2 (Addgene #12260), using 72 μl of lipofectamine 2000 in 3 ml of optiMEM medium. Opti-MEM was added to a final volume of 8 ml, containing 25 μM chloroquine, 1 × L-glutamine, 1 × non-essential amino acids, 1 × sodium pyruvate and 10% foetal bovine serum. The transfection medium was replaced the next day with 10 ml Opti-MEM supplemented with 1 × L-glutamine, 1 × non-essential amino acids and 1 × sodium pyruvate. Lentiviral supernatant was harvested 24 hours later, filtered through a 0.4 μm syringe filter and stored as 1 ml aliquots at −80 °C.
Cell transduction
GBM cell lines were transduced with lentivirus in matrigel-coated 6-well tissue culture plates in 4 ml of RHB-A medium by adding 1 ml of unconcentrated lentivirus and 1 μg/ml polybrene followed by centrifugation at room temperature for 45 minutes at 440 g. Virus-containing medium was replaced next day with fresh RHB-A medium and antibiotic selection commenced 48 hours after lentiviral transduction began.
GBM xenograft models
GBM xenograft models were initiated in 5-week old NOD/SCID mice housed under pathogen-free conditions. For intracranial cell injections 150,000 cells in 2 μl of 10 ng/ml laminin in PBS were injected 3 mm below the brain surface, 1.6 mm rostral of the bregma and 0.8 mm right of the midline, using a 25G Hamilton needle and a 2 μl syringe in a stereotaxic frame. Mice were anaesthetised with 2% isoflurane (Abbott) in oxygen at a flow rate of 0.8 litres per minute and given 100 μg of carprofen (Pfizer) subcutaneously for analgesia. The burr hole through which the cells were injected was sealed with bone wax and the midline scalp incision was closed with Vetbond (3M) tissue adhesive. Mice were euthanased when they exhibited signs of significant morbidity (hunching, weight loss, rough coat, ataxia, head tilt, paralysis). All studies were conducted according to protocols approved by the Animal Ethics Committee of the QIMR Berghofer Medical Research Institute. The “Principles of laboratory animal care” (NIH publication No. 86–23, revised 1985) were followed as well as the “Australian code of practice for the care and use of animals for scientific purposes”, 8th edition 2013 and the “Queensland Animal Care and Protection Act 2001”.
Bioluminescence imaging of xenograft tumours
Bioluminescence imaging of intracranial xenograft tumours was performed 5 minutes after injecting 0.5 mg of luciferin in 100 μl of DPBS (Dulbecco’s Phosphate-Buffered Saline, without Mg2+ and Ca2+) intraperitoneally into isoflurane-anaesthetised NOD/SCID mice using an IVIS Spectrum In Vivo Imaging System (Perkin Elmer).
Exome sequencing
Genomic DNA was prepared from cultured cells using a QIAmp DNA mini kit (Qiagen) with RNase A treatment. Exome sequencing was performed by the Beijing Genomics Institute (BGI) using Truseq (BAH1 and WK1) or SureSelect (the remainder) exon capture and Illumina HiSeq 2000 technology. Data filtering, read alignment, SNV calling, annotation and statistics were performed by BGI.
Copy number determination
Where investigated, gene copy number was determined by inspection of exon reads using Integrative Genomics Viewer51,52.
Microarray analysis of gene expression
Microarray analysis of gene expression was performed using HumanHT-12_v4_BeadChip arrays (Illumina). RNA was extracted from patient tumour tissue, adherent cell cultures and xenograft tumours using RNeasy mini kits (Qiagen). Biotinylated cRNA was prepared using TotalPrep RNA amplification kits (Ambion). Microarray hybridisation and Cy3-streptavidin labelling was performed according to recommended protocols (Illumina). BeadChips were imaged with an iScan beadchip reader (Illumina) running BeadStudio software (Illumina). Raw data was exported to GenomeStudio (Agilent) where it was quantile normalised and background subtracted. Illumina beadchips allow a detection P value to be calculated as an estimate of gene measurements relative to background. P < 0.05 for all samples on a beadchip for a given probe was used as a cutoff to compile gene expression datasets. Where genes were represented by multiple probes, the highest value probe was used for gene-level summaries (tumour 11,267 genes; cell lines 10,435 genes; xenografts 7,079 genes). Illumina idat files are available for download (qimrberghofer.edu.au/q-cell/) and GenomeStudio-processed probe-level datasets (Supplementary Data S3) are provided for independent validation of our findings and as a reference for gene expression measurement.
GBM molecular subtyping
GBM molecular subtyping was performed by hierarchical cluster analysis using MeV (http://mev.tm4.org) with gene sets reported for proneural, classical and mesenchymal GBM4, a Pearson distance metric and average linkage criteria, and also by gene set enrichment analysis53,54 (GSEAv2.2.4) in the pre-ranked mode using the proneural, classical and mesenchymal.rnk files in the Molecular Signatures Database v6.0. Gene ranking was according to z-scores for RNA expression determined by microarray analysis. Heatmaps were created using Excel.
RNAseq
RNA was extracted from frozen cell pellets using a Qiagen AllPrep DNA/RNA Minikit (Cat No: 80204). RNA integrity was assessed using an Agilent RNA 6000 Eukaryote Total RNA kit (Cat. No: 5067–1511) and Bioanalyzer 2100 instrument. All RNA samples had an RNA Integrity Number (RIN) >9.0. RNA libraries were prepared using an Illumina TruSeq Stranded mRNA Library Preparation Kit Set A (Cat. No: RS-122-2101). RNA libraries were sequenced using an Illumina NexSeq® 500/550 High Output Kit v2 (150 cycles) (Cat. No: FC-404-2002) and NexSeq® 550 instrument.
Paired-end fastq files had adapters trimmed and flanking “N” bases removed for each paired-end read using Cutadapt v1.18. Trimmed files were aligned to the human GRCh37_ICGC_standard_v2 reference using STAR v2.5.2a. Quality control was performed using RNA-SeQC v1.1.8. Expected read counts and FPKM were calculated using RSEM v1.2.30, with a forward probability of 0 as the reads were stranded and derived from reverse strand. Finally, expected counts and FPKM for both genes and isoforms, were matched to gene/transcript annotation information using in-house scripts to provide expected counts and FPKM summary files for each sample.
Western blotting
Cells (~70–90% confluent, passage 15) were washed 3 × in ice-cold PBS, then lysed on ice with ice-cold RIPA buffer (500 μl per plate, ThermoFisher Scientific) containing cOmpleteTM, Mini, EDTA-free Protease Inhibitor Cocktail (Merck) and PhosSTOP (Merck). Cells were disrupted by repeated passage through a 27G needle and crude lysates centrifuged at 20,000 g for 15 mins at 4 °C. Supernatants were collected, and protein content determined by Bio-Rad Protein Assay. 80 μg of total protein was loaded for each cell line per lane and proteins separated on 8–16% CriterionTM TGXTM Precast Midi Protein Gels (12 + 2 well, 45 μl, Bio-Rad) and transferred onto Immun-Blot® PVDF Membrane (Bio-Rad). Membranes were blocked in 5% BSA/TBST, washed in TBST, and protein levels determined using the following antibodies: EGFR (Abcam, ab52894, 1:1000), c-myc (CST, #5605, 1:1000), PTEN (CST, #9188, 1:1000) and β-actin (CST, #3700, 1:3000).
Proteomics
GBM cell lines were grown as tumourspheres for three days. Cells were lysed in buffer containing 2% SDS, 100 mM Tris pH 7.6, protease and phosphatase inhibitors followed by precipitation with acetone. The protein precipitate was resuspended in 8 M urea, followed by reduction with DTT, alkylation with iodoacetamide and digested with trypsin using a standard protocol (https://www.nature.com/articles/ni.3693). Peptides were desalted and separated on a C18 analytical column (Waters) interfaced to an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). All raw files were searched against human proteome sequences from UniProt using the based label-free quantification algorithm in MaxQuant (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4159666/) (protein and peptide FDR = 0.01). Proteins of interest were extracted manually from the total list.
Flow cytometry
Flow cytometry was performed with an LSR Fortessa (BD Biosciences) for data acquisition and fluorochrome-conjugated antibodies targeting CD133 (Miltenyi Biotec, AC133, 1:20), CD15 (BD Biosciences, 1:100), CD44 (in-house Hermes 3, 1:500), CD49f (Jomar Biosciences, 14-0495, 1:100), EGFR (in-house, M528, 0.5 μg/ml), EGFRvIII (in-house, 806, 0.5 μg/ml), PDGFRα (R&D Systems, MAB1264, 0.5 μg/ml), EphA2 (in-house, 1F7, 0.5 μg/ml), EphA3 (in-house, IIIA4, 0.5 μg/ml), EphA4 (in-house, 1F9, 0.5 μg/ml), FGFR1 (Abcam, ab823, 1:100), FGFR2 (Abcam, ab89476, 1:100), FGFR3 (R&D Systems, MAB766, 0.5 μg/ml), c-Met (R&D Systems, MAB3582, 0.5 μg/ml), Sox2 (R&D Systems, MAB2018, 0.5 μg/ml), Nestin (R&D Systems, MAB1259, 0.5 μg/ml), GFAP (Dako, Z0334, 1:100), β-III tubulin (Promega, G7121, 1:100) and MBP (Sigma Aldrich, M3821, 1:100). Cells were incubated with antibody on ice for 30 minutes in PBS containing 2% FBS. Where required, cells were fixed and permeabilised using Intracellular Fixation & Permeabilization Buffer (eBioscience) and stained with antibodies in 1 × Permeabilization Buffer. Data analysis was performed using Flowjo (Treestar) software.
Statistical analysis
Unless otherwise noted, data are presented as the mean and standard error of the mean. GSEA reports normalised enrichment scores with the associated false discovery rate and familywise-error rate. P < 0.05 was considered statistically significant.
Materials availability
All GBM cell lines are available to research groups with a standard materials transfer agreement and can be obtained by contacting the corresponding author or via the website, qimrberghofer.edu.au/q-cell/. Cell lines (not expressing luciferase) will be distributed from passage 10 onwards. Lentiviral plasmids can be obtained from Addgene at https://www.addgene.org/Brett_Stringer/.
Supplementary information
Acknowledgements
We thank the QIMR Berghofer Medical Research Institute (QIMR-B) Flow Cytometry, Histology, DNA Sequencing and Animal Facility staff for their excellent technical support, Mitchell Stark, Ross Koufariotis, Pamela Mukhopadhyay and Nic Waddell for bioinformatics assistance, Glen Boyle for assistance with microarrays and the QIMR-B External Relations, Information Technology, Legal and Business Development teams for website development. BWS thanks Prof Mark Smyth, Head of the Immunology in Cancer and Infection Laboratory in whose laboratory some of the experiments were performed and Dr Liam Town from the Immunology in Cancer and Infection Laboratory for his practical assistance. This work was supported by a Private Practice Foundation research grant from the Royal Brisbane and Women’s Hospital and a Rio Tinto Ride to Conquer Cancer research grant from the QIMR Berghofer Medical Research Institute to BWS, BWD and AWB and an NHMRC project grant (APP1028552) to AWB and TGJ. BWS, BWD, TGJ and AWB are members of the Brain Cancer Discovery Collaborative, which is supported by the Cure Brain Cancer Foundation.
Author Contributions
B.W.S. and B.W.D. conceived and coordinated the project with feedback from A.W.B. and T.G.J. P.I., S.A., P.L., G.T., S.C., C.W. and R.L.J. obtained patient consent for donated tissue. P.L., G.T., S.C., C.W. and R.L.J. acquired tumour tissue during surgery. B.W.S., B.W.D., P.R.J., K.S.E., Z.C.B., Y.C.L. and K.G. collected patient tissue following surgery and established the cell lines. B.W.S. designed and constructed the lentiviral plasmids. B.W.S. and P.R.J. determined cell line antibiotic sensitivities and established bioluminescent cell lines. K.E.S. and Z.C.B. performed intracranial injections. K.E.S., Z.C.B. and K.G. monitored animal welfare. B.W.S. performed proliferation assays, determined IDH1 mutation status, arranged exome sequencing, STR analysis and mycoplasma testing. B.W.S. performed the microarray experiments and GBM subtype determination. S.A. performed RNAseq. R.C.J.D. conducted proteomic experiments. C.O. performed western blotting. Z.C.B. performed flow cytometry. H.D. and A.D. were responsible for pyrosequencing. T.G.J. provided anti-EGFR and anti-EGFRvIII monoclonal antibodies. B.W.S. analysed the data and prepared the figures. B.W.S. wrote the manuscript with feedback from A.W.B. and T.G.J. A.W.B. and T.G.J. provided funding. All authors read and approved the final submission.
Data Availability
The datasets generated during the current study are available for download at qimrberghofer.edu.au/q-cell/ and/or as Supplementary Data S1–6. Microarray data have been deposited with the GEO repository with the series accession number GSE118793. RNAseq raw data is available for download from the NCBI sequence read archive with SRA accession number PRJNA508446.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Brett W. Stringer and Bryan W. Day contributed equally.
Terrance G. Johns and Andrew W. Boyd jointly supervised this work.
Change history
1/21/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41277-z.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SS, Pirollo KF, Chang EH. Isolation and Culturing of Glioma Cancer Stem Cells. Curr Protoc Cell Biol. 2015;67:23 10 21–10. doi: 10.1002/0471143030.cb2310s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Fael Al-Mayhani TM, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day BW, et al. Glioma surgical aspirate: a viable source of tumor tissue for experimental research. Cancers (Basel) 2013;5:357–371. doi: 10.3390/cancers5020357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shihab HA, et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan C, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos B, et al. Expression and regulation of AC133 and CD133 in glioblastoma. Glia. 2011;59:1974–1986. doi: 10.1002/glia.21239. [DOI] [PubMed] [Google Scholar]
- 24.Binda E, et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day BW, et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell. 2013;23:238–248. doi: 10.1016/j.ccr.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Xie Y, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2:1351–1363. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2017;32:42–56 e46. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhat KPL, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci USA. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flavahan WA, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stringer BW, et al. Nuclear factor one B (NFIB) encodes a subtype-specific tumour suppressor in glioblastoma. Oncotarget. 2016;7:29306–29320. doi: 10.18632/oncotarget.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenall SA, et al. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis. 2017;6:e336. doi: 10.1038/oncsis.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abuhusain HJ, et al. A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J Biol Chem. 2013;288:37355–37364. doi: 10.1074/jbc.M113.494740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tivnan A, et al. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumour Biol. 2014;35:1459–1465. doi: 10.1007/s13277-013-1200-6. [DOI] [PubMed] [Google Scholar]
- 35.Lim YC, et al. Increased sensitivity to ionizing radiation by targeting the homologous recombination pathway in glioma initiating cells. Mol Oncol. 2014;8:1603–1615. doi: 10.1016/j.molonc.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rundle-Thiele D, et al. Using the apparent diffusion coefficient to identifying MGMT promoter methylation status early in glioblastoma: importance of analytical method. J Med Radiat Sci. 2015;62:92–98. doi: 10.1002/jmrs.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puttick S, et al. EphA2 as a Diagnostic Imaging Target in Glioblastoma: A Positron Emission Tomography/Magnetic Resonance Imaging Study. Mol Imaging. 2015;14:385–399. [PubMed] [Google Scholar]
- 38.Grundy TJ, et al. Differential response of patient-derived primary glioblastoma cells to environmental stiffness. Sci Rep. 2016;6:23353. doi: 10.1038/srep23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosein AN, et al. The effect of valproic acid in combination with irradiation and temozolomide on primary human glioblastoma cells. J Neurooncol. 2015;122:263–271. doi: 10.1007/s11060-014-1713-x. [DOI] [PubMed] [Google Scholar]
- 40.Phoa AF, et al. Pharmacology of novel small-molecule tubulin inhibitors in glioblastoma cells with enhanced EGFR signalling. Biochem Pharmacol. 2015;98:587–601. doi: 10.1016/j.bcp.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Zakaria Z, et al. Patient-derived glioblastoma cells show significant heterogeneity in treatment responses to the inhibitor-of-apoptosis-protein antagonist birinapant. Br J Cancer. 2016;114:188–198. doi: 10.1038/bjc.2015.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tivnan A, et al. Anti-GD2-ch14.18/CHO coated nanoparticles mediate glioblastoma (GBM)-specific delivery of the aromatase inhibitor, Letrozole, reducing proliferation, migration and chemoresistance in patient-derived GBM tumor cells. Oncotarget. 2017;8:16605–16620. doi: 10.18632/oncotarget.15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q, et al. Structural Optimization and Pharmacological Evaluation of Inhibitors Targeting Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases (DYRK) and CDC-like kinases (CLK) in Glioblastoma. J Med Chem. 2017;60:2052–2070. doi: 10.1021/acs.jmedchem.6b01840. [DOI] [PubMed] [Google Scholar]
- 44.Dobber A, et al. Development and Biological Evaluation of a Photoactivatable Small Molecule Microtubule-Targeting Agent. ACS Med Chem Lett. 2017;8:395–400. doi: 10.1021/acsmedchemlett.6b00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilabert-Oriol, R. et al. Dianthin-30 or gelonin versus monomethyl auristatin E, each configured with an anti-calcitonin receptor antibody, are differentially potent in vitro in high-grade glioma cell lines derived from glioblastoma. Cancer Immunol Immunother (2017). [DOI] [PMC free article] [PubMed]
- 46.Mikeska T, Felsberg J, Hewitt CA, Dobrovic A. Analysing DNA methylation using bisulphite pyrosequencing. Methods Mol Biol. 2011;791:33–53. doi: 10.1007/978-1-61779-316-5_4. [DOI] [PubMed] [Google Scholar]
- 47.Callus, B. A. et al. Cytoplasmic p53 is not required for PUMA-induced apoptosis. Cell Death Differ15, 213–215 author reply 215-216 (2008). [DOI] [PubMed]
- 48.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available for download at qimrberghofer.edu.au/q-cell/ and/or as Supplementary Data S1–6. Microarray data have been deposited with the GEO repository with the series accession number GSE118793. RNAseq raw data is available for download from the NCBI sequence read archive with SRA accession number PRJNA508446.