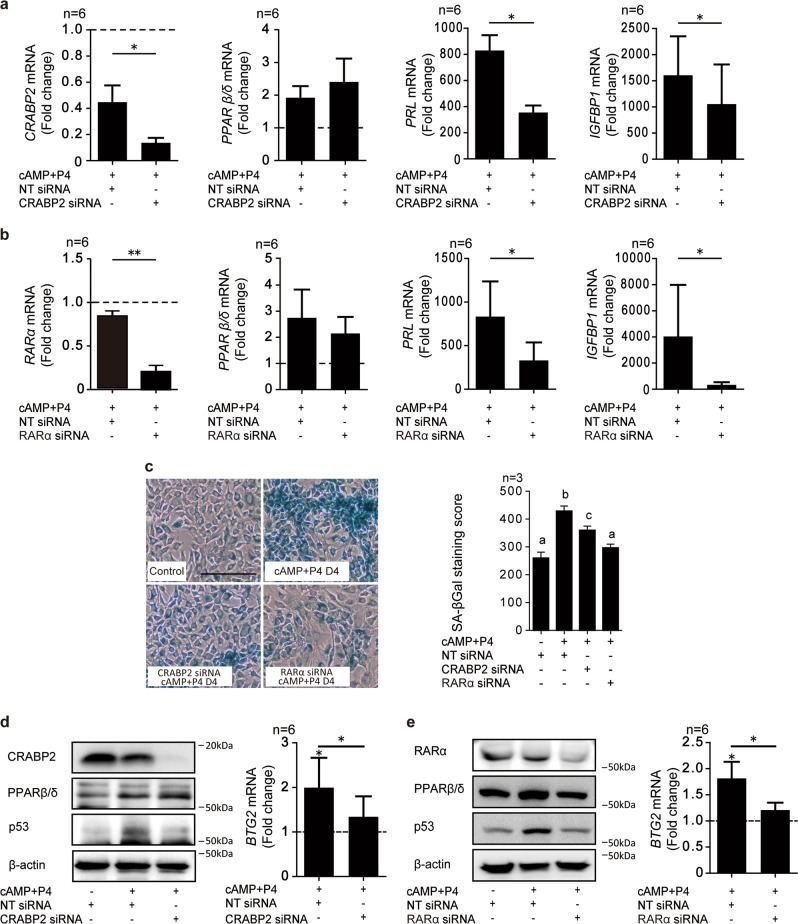
Fig. 5. CRABP2 or RARα knockdown inhibits decidualization.
a, b RTQ-PCR analysis of CRABP2, RARα, PPARβ/δ, PRL, and IGFBP1 transcript levels in 6 independent HESC cultures first transfected with NT, CRABP2 (a) or RARα (b) siRNA and then treated with cAMP and P4 for 4 days. Data show fold-change (mean ± SEM) relative to vehicle control (dotted line). *P < 0.05; **P < 0.01. c Representative SAβG staining of 3 independent HESC cultures first transfected with NT, CRABP2, or RARα siRNAs and then decidualized for 4 days. Original magnification: ×100. Scale bar: 200 μm. Data shows SAβG staining intensity score resulted from 300 decidual cells in 3 random fields Different letters above error bars denote significance at P < 0.05 (right panel). d, e Representative Western blot of RA-related gene proteins and p53 in whole-cell lysates obtained from 6 independent HESC cultures treated as indicated. β-actin serves as a loading control (left panel). RTQ-PCR analysis of BTG2 transcript levels in primary parallel cultures (right panel). The results show the fold-change (mean ± SEM)