 Genome mining and heterologous expression of an orphan cluster led to the discovery of ashimides featuring an unusual cyclization mechanism.
Genome mining and heterologous expression of an orphan cluster led to the discovery of ashimides featuring an unusual cyclization mechanism.
Abstract
The ever-increasing bacterial genomic repositories reveal a great number of uncharacterized biosynthetic gene clusters, representing a tremendous resource for natural product discovery. Genome mining of the marine Streptomyces sp. NA03103 indicates the presence of an orphan nonribosomal peptide synthetase (NRPS) gene cluster (asm), to which there are no homologous gene clusters in the public genome databases. Heterologous expression of the asm gene cluster in the S. lividans SBT18 strain led to the discovery of two novel cyclopeptides, ashimides A and B (1 and 2), with 2 showing cytotoxic activity. In addition, we use bioinformatic analysis, gene inactivation and stable isotope labelling experiments, as well as in vitro biochemical assays, to present a coherent and novel assembly line for ashimide biosynthesis, featuring an unusual desaturation, halogenation and cyclization cascade catalyzed by a P450 monooxygenase and a FAD-dependent halogenase.
Nonribosomal peptides (NRPs) produced by actinobacteria constitute a large group of secondary metabolites with potent biological activities as exemplified by vancomycin, dactinomycin, and daptomycin. Nature developed modular biosynthetic machinery to synthesize NRPs, in which each module in the nonribosomal peptide synthetase (NRPS) assembly line activates and incorporates an amino acid into a growing peptide.1–5 According to this rule, the amino acid building block composition of NRPs can correlate well with their corresponding NRPS architectures.6,7 With the massive accumulation of bacterial genome data, it has become obvious that the biosynthetic potential of NRPs greatly exceeds the number of compounds that have been isolated.8,9 Orphan gene clusters, which are gene loci to which no known homologous gene clusters can be found, represent a particularly rich and untapped source for novel natural product discovery. Genome mining targeting the orphan NRPS gene cluster is therefore a powerful approach to discover novel NRPs.
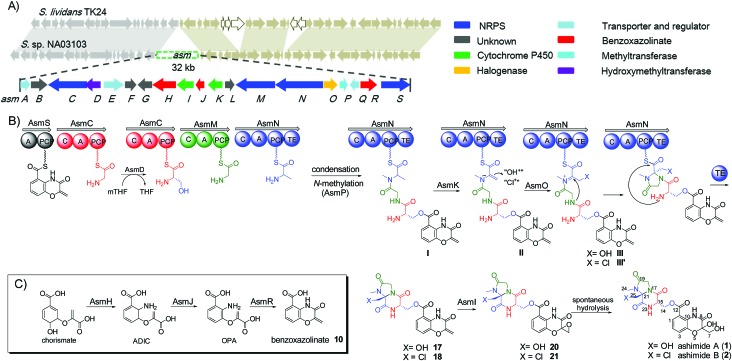
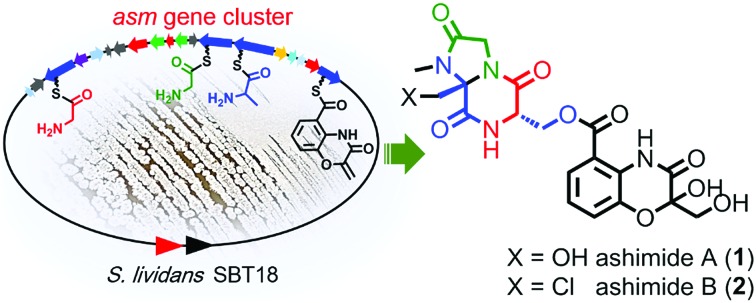
During our genome survey of sequenced strains, an orphan NRPS gene cluster (asm) was identified from a marine Streptomyces sp. NA03103. The asm gene cluster is unusual for three reasons: (i) no homologous gene clusters can be found in the public genome databases through ClusterBlast, a dereplication module in online software antiSMASH;10 (ii) the upstream and downstream genes flanking the putative asm gene cluster are highly conserved (>93% identity) in a model Streptomyces strain S. lividans TK24 (Scheme 1); (iii) all four NRPSs (AsmC, AsmM, AsmN and AsmS) are small monomodule NRPSs, which are in sharp contrast to the canonical multimodule NRPS megaenzymes in bacterial gene clusters.11 The putative asm gene cluster embedded in the conserved region is proposed to only span ∼32 kb and consists of 19 genes (asmA–asmS) (Scheme 1 and Table S3†). In addition, several notable tailoring enzymes are also present including an FAD-dependent halogenase (asmO), two P450 monooxygenases (asmI and asmK), and a methyltransferase gene (asmP).
Scheme 1. Biosynthesis of ashimides. (A) The asm gene cluster and schematic comparison of asm neighbouring genes in S. sp. NA03103 with their homologs in S. lividans TK24; (B) proposed biosynthetic pathway of ashimides A and B; (C) proposed biosynthetic pathway of the benzoxazolinate moiety in ashimides. mTHF, 5,10-methylenetetrahydrofolate; THF, tetrahydrofolate; ADIC, 2-amino-2-desoxyisochorismate; OPA, 3-enolpyruvoylanthranilate.
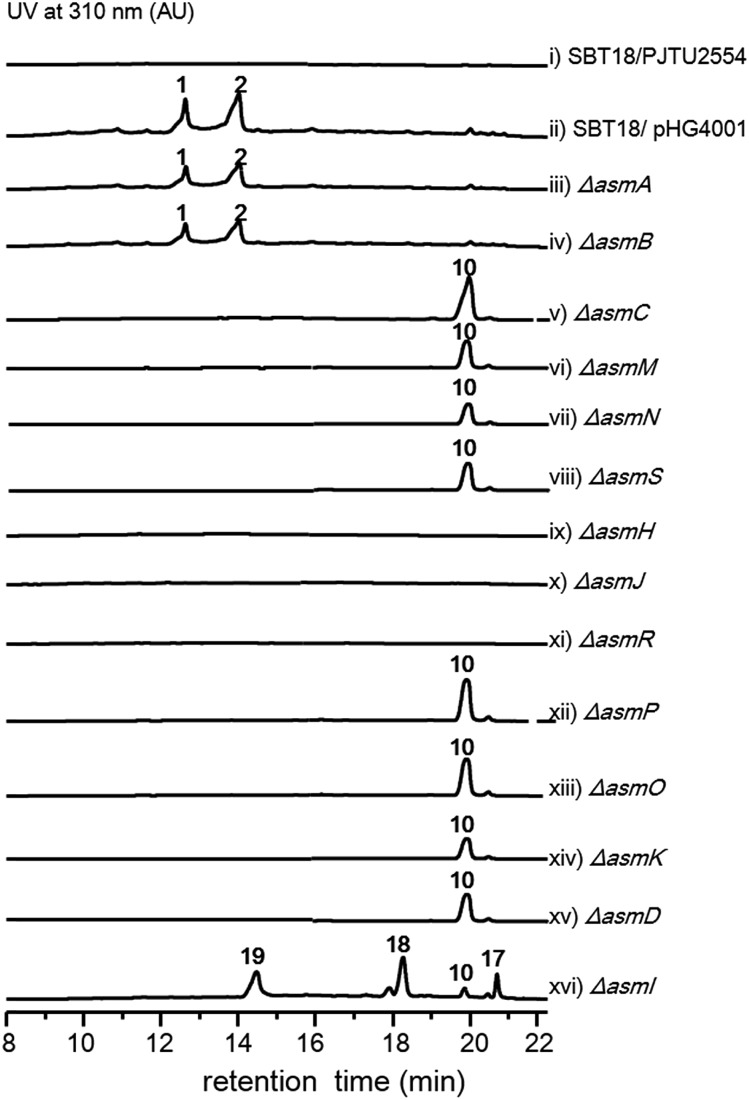
With the gene cluster in hand, we first aimed to identify its encoded natural products in strain NA03103. Somewhat to our surprise, no corresponding compounds could be isolated from metabolic extracts of this wild-type strain, even when cultured under a wide variety of growth conditions. Putatively, this gene cluster is silent in this strain under current laboratory conditions. To bypass the endogenous regulatory control and activate the cryptic pathway, we sought to clone the intact asm gene cluster into a heterologous host. Thus, we constructed a cosmid library of S. sp. NA03103 using a Streptomyces integrative vector pJTU2554 according to a standard procedure.12 One cosmid, pHG4001 (Fig. S1†), containing the full length of the asm gene cluster was obtained and transferred into S. lividans SBT18 for heterologous expression. Metabolic extracts from the resulting S. lividans SBT18/pHG4001 strain were obtained and analyzed by HPLC and LC/MS. In comparison to the S. lividans harboring an empty cosmid vector, S. lividans SBT18/pHG4001 produced two major peaks (1 and 2) with molecular weights of 464 and 482 (Fig. 1), respectively. It is of note that 2 showed a characteristic ratio of monochlorinated isotopic signals ([M + Na]+ : [M + Na + 2]+ = 3 : 1) (Fig. S2†), in line with the presence of a putative FAD-dependent chlorinase (AsmO) encoded in the gene cluster. Hence, we carried out a large-scale (20 L) fermentation from which we isolated compounds 1–9 (Fig. S3†), the latter seven of which were barely observed in the HPLC profile.
Fig. 1. HPLC analysis of metabolite extracts from mutant strains.
Compound 1 (1.0 mg L–1) was obtained as a pale yellow crystal with a molecular formula of C19H20N4O10 as determined by its HRESIMS and 13C NMR data (Table S4†). The planar structure, along with the absolute configurations, was unambiguously determined by analyzing its NMR and single-crystal X-ray diffraction data with a flack parameter value of –0.03(7) collected using Cu Kα at low temperature (Fig. S4†). Compound 2 (1.3 mg L–1) has a molecular formula of C19H19ClN4O9. The 1H and 13C NMR spectra highly resembled those of 1 except for the absence of the proton signal for 25-OH and a chemical shift value for C-25 (δC 47.3 for 2 and 61.4 for 1), which indicated that the OH group in C-25 was substituted by a chlorine atom. Further interpretation of 1D, 2D NMR and CD data established the structure of 2 (Table S5, Fig. S5†). Structures of 3–9 were characterized through extensive analyses of HRMS and NMR data (Tables S6–S12†). To assess the biological activity, we then evaluated the cytotoxicity of 1–9 against MCF-7 cell lines using the MTT assay. Only 2 exhibited cytotoxicity with an IC50 value of 19.8 μM, whereas others are inactive. The flow cytometry analysis showed that 2 could induce apoptosis in a dose-dependent manner in MCF-7 cells (Fig. S6†).
The two most abundant compounds, ashimides A and B (1 and 2), contain two rare structural features including the 5/6 bicyclic imidazo[1,2-a]diketopiperazine and benzoxazolinate moieties, the former of which has never been observed among natural products, hinting that a novel cyclization strategy may be involved in the biosynthesis of ashimides. Additionally, the benzoxazolinate moiety, which plays a crucial role in intercalating double-stranded DNA,13 has only been reported in enediyne antitumor agents C-1027 and auromomycin.14,15 These unusual structural features encouraged us to investigate the biosynthetic machinery of ashimides in depth. For this purpose, we systematically inactivated genes in the asm cluster through in-frame deletion (Fig. S7†), which was achieved through homologous recombination between the cosmid pHG4001 and a shuttle vector carrying the mutation (see the ESI†).16 The resultant mutant strains were fermented, with the S. lividans SBT18/pHG4001 strain as a control, to investigate the effect of these genes on 1 and 2 biosynthesis.
Deletion of two boundary genes asmA and asmB, which are annotated to be a response regulator and a histidine kinase, respectively, did not affect the production of 1 and 2 (Fig. 1 and S8†). Inactivation of NRPS genes, asmC, asmM, asmN or asmS, as expected no longer produced 1 and 2 but instead clearly accumulated a new compound 10 (Fig. 1 and S8†). Isolation of 10 from the ΔasmC strain followed by NMR characterization (Table S13†) indicated that 10 is the benzoxazolinate compound (Scheme 1C). We proposed that the formation of 10 takes place prior to adenylation and the loading module AsmS may directly accept 10 as a substrate.
AsmH shares high sequence homology (52% identity) with PhzE, a biochemically characterized 2-amino-2-desoxyisochorismate (ADIC) synthase in the first step of phenazine biosynthesis,17 whereas AsmJ and AsmR are homologous to SgcG (48% identity) and SgcD5 (48% identity),18 respectively, two tandem enzymes involved in the conversion of ADIC to 10 in the biosynthesis of a benzoxazolinate motif of C-1027.14 Therefore, these three enzymes are most likely sufficient for the biosynthesis of 10 (Scheme 1). Knocking out asmH, asmJ or asmR abolished the production of 1 and 2, but did not accumulate any observed peaks (Fig. 1 and S8†). The production of 1 and 2 can be restored when feeding 10 into ΔasmH, ΔasmJ or ΔasmR mutants individually (Fig. S9†), confirming their involvement in the biosynthesis of 10 and also indicating that 10 is an on-pathway intermediate.
Deletion of the asmP gene, which encoded a methyltransferase (Table S3†), resulted in the complete abolition of 1 and 2 with only the accumulation of 10 (Fig. 1 and S8†). This indicates that N-methylation is essential for ashimide biosynthesis. In addition, supplementation of the putative N-methylated amino acid precursors including 3-chloro-N-methylalanine (11), N-methylserine (12), or N-methylalanine (13) into ΔasmP could not restore the production of 1 or 2 (Fig. S9†), suggesting that the methylation step most likely takes place after amino acid binding to the NRPS.19,20 A similar scenario has also been observed in α-lipomycin biosynthesis.21
The substituted groups at C-25 of 1 and 2 are hydroxyl and chlorine, respectively, raising the question of their origin. Knocking out the FAD-dependent halogenase gene, asmO, eliminated the production of 1 and 2, and accumulated 10, excluding the possibility that 1 is the precursor of 2, and vice versa. As the FAD-dependent halogenase/hydroxylase requires electron rich alkenes (such as phenols, pyrroles, or indoles) for halogenation/hydroxylation,22 a putative alkene-containing intermediate is proposed to form during biosynthesis. In accordance with this hypothesis, a P450 monooxygenase AsmK, which shows moderate sequence homology (34% identity) to LtmK, a desaturase in lactimidomycin biosynthesis,23 is encoded in the asm gene cluster. The mutant strain ΔasmK only accumulated 10 (Fig. 1 and S8†). Feeding of 11, 13 and 3-chloroalanine (14) to ΔasmO cannot restore the production of 1 or 2 (Fig. S9†). Therefore, we reasoned that the AsmN may activate and accept alanine as a substrate instead of serine. After elongation, the alanine building block is desaturated by AsmK to form a 2-aminoacrylic acid containing intermediate. AsmO may play a bifunctional role in introducing either a hydroxyl or a chlorine group at the newly generated alkene position (Scheme 1), as the FAD-dependent halogenases can be viewed as variants of FAD-dependent hydroxylases.22
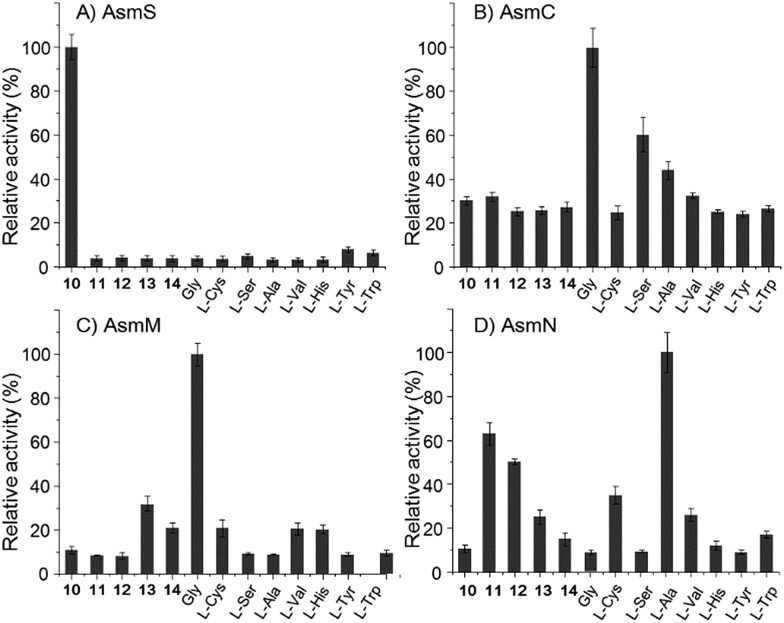
However, the A domain substrate specificity of AsmN predicted by NRPSpredictor2 is cysteine instead of alanine as hypothesized above (Table S20†).24 In addition, A domains in AsmC, AsmM, and AsmS were predicted to incorporate cysteine, glycine, and glycine/alanine/valine, respectively (Table S20†),24 which is not in full agreement with amino acid composition in ashimides. To determine the substrate specificity of the A domains, asmC, asmM, asmN, and asmS genes were individually overexpressed from Escherichia coli BL21(DE3), and the full-length NRPS proteins were purified (Fig. S10†). The adenylation activity towards various amino acid substrates was evaluated by monitoring pyrophosphate production through a fluorogenic pyrophosphate sensor.25 As shown in Fig. 2, AsmS specifically accepts benzoxazolinate (10) as the substrate, which is consistent with the structures of 1 and 2, as 10 should be the first amino acid to be incorporated. Interestingly, both AsmC and AsmM showed clear preferences for glycine, although substrate specificity of AsmC was relatively broad. The last module, AsmN, prefers l-alanine as the substrate over 3-chloro-N-methylalanine (11), N-methylserine (12), N-methylalanine (13), 3-chloroalanine (14) or cysteine. These findings, in conjunction with the results of the in vivo knocking out experiments, strongly suggested that benzoxazolinate, glycine, glycine and l-alanine are the substrates of AsmS, AsmC, AsmM and AsmN, respectively, in ashimide biosynthesis.
Fig. 2. The relative adenylation activities for (A) AsmS, (B) AsmC, (C) AsmM, and (D) AsmN. Error bars represent SEM (n = 3).
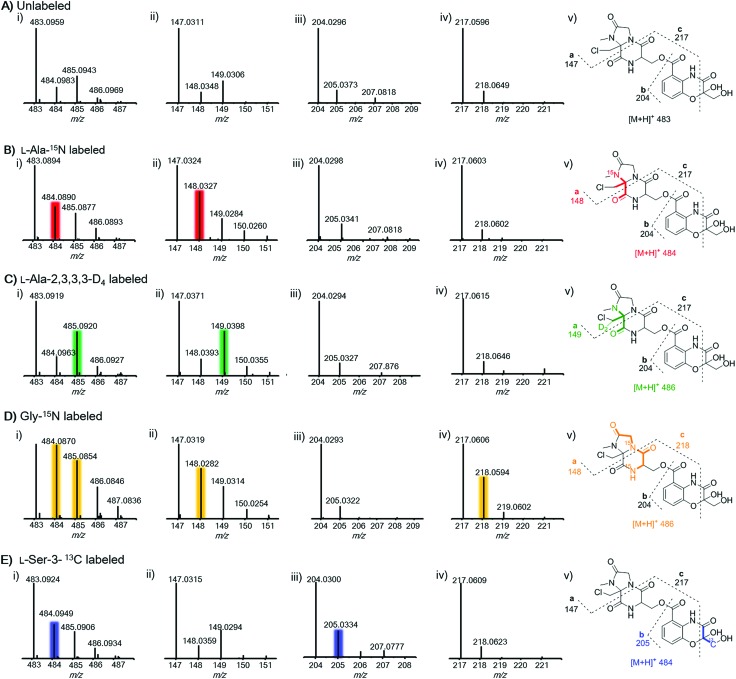
To further prove this, we next probed the origin of amino acid building blocks by feeding a stable isotope labeled amino acid to the culture. In contrast to the unlabeled ion (Fig. 3A(i)), LC-MS analysis of 15N-labelled alanine clearly showed the increment of the ion peak at m/z 484.1 (2, [M + H]+) (Fig. 3B(i)), indicating that one alanine was incorporated into 2. To ascertain which N atom was labeled by 15N, 2 L scale fermentation was conducted with feeding labeled 15N-alanine, leading to the purification of 0.2 mg of 2. The 15N–1H HMBC spectrum clearly indicated that H-24 and H-25 showed correlations with N-20 (Fig. S11†), revealing that the 3-chloroalanine building block was indeed derived from alanine, which was reinforced by the observed MS/MS fragmentation of 148.0 (Fig. 3B(ii)). Supplementation of alanine-D4 into the culture led to +2 Dalton mass shifts in 2 (Fig. 3C(i)) and a chlorine-containing fragment (m/z 149.0) (Fig. 3C(ii)), further supporting the fact that 3-chloro-alanine originated from alanine. When the culture was supplemented with 15N-glycine, a +2 Dalton mass shift was observed (Fig. 3D(i)), indicating that two molecules of glycine were incorporated into 2. The relative abundance of ion peaks at m/z 148.0 (Fig. 3D(ii)) and 218.1 (Fig. 3D(iv)) in MS/MS analysis revealed that two glycines were incorporated in the second and third NRPS modules, respectively. To further verify if serine can be incorporated into 2, 13C3-serine was added to the culture. The MS and MS/MS ion peaks with a +1 Dalton mass shift were observed only for the benzoxazolinate-containing part (Fig. 3E(iii)). This can be explained by the fact that the three-carbon unit in the benzoxazolinate moiety is derived directly from phosphoenolpyruvic acid (PEP), which originates from serine through the pyruvate intermediate (Fig. S12†).26
Fig. 3. MS and MS2 fragmentation analysis of 2 when targeting the unlabeled (panel A) and after feeding with labeled amino acids including l-alanine-15N (panel B), l-alanine-2,3,3,3-D4 (panel C), glycine-15N (panel D), and l-serine-3-13C (panel E). Panel (A): parent MS signal for unlabeled 2 (i, m/z 483, [M + H]+); MS2 ion signals for fragments of a (ii, m/z 147), b (iii, m/z 204), and c (iv, m/z 217); (v) MS2 fragmentation ion pattern for unlabeled 2. Panel (B): parent MS signal for 2 after feeding l-alanine-15N (i, m/z 484, [M + H]+); MS2 ion signals for fragments of a (ii, m/z 148), b (iii, m/z 204), and c (iv, m/z 217); (v) MS2 fragmentation ion pattern for 2 after feeding l-alanine-15N. Panel (C): parent MS signal for 2 after feeding l-alanine-2,3,3,3-D4 (i, m/z 484, [M + H]+); MS2 ion signals for fragments of a (ii, m/z 148), b (iii, m/z 204), and c (iv, m/z 217); (v) MS2 fragmentation ion pattern for 2 after feeding l-alanine-2,3,3,3-D4. Panel (D): parent MS signal for 2 after feeding glycine-15N (i, m/z 485, [M + H]+); MS2 ion signals for fragments of a (ii, m/z 148), b (iii, m/z 204), and c (iv, m/z 218); (v) MS2 fragmentation ion pattern for 2 after feeding glycine-5N. Panel (E): parent MS signal for 2 after feeding l-serine-3-13C (i, m/z 484, [M + H]+); MS2 ion signals for fragments of a (ii, m/z 147), b (iii, m/z 205), and c (iv, m/z 217); (v) MS2 fragmentation ion pattern for 2 after feeding l-serine-3-13C.
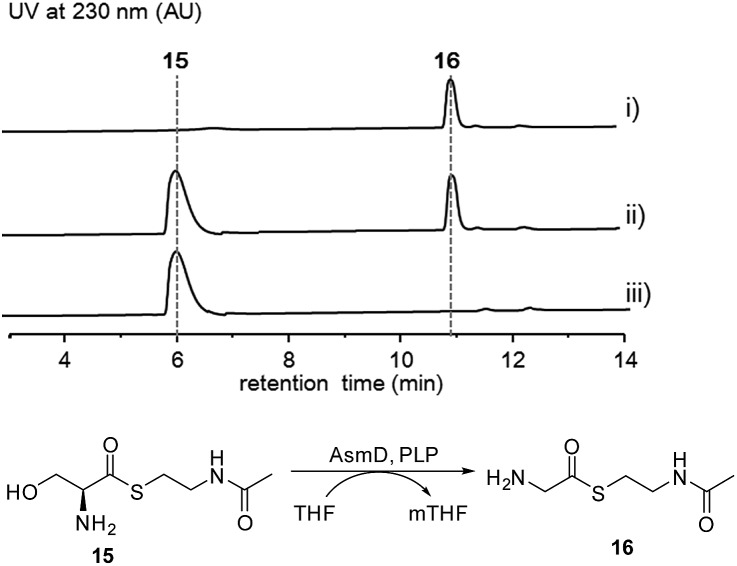
Thus, both the A domain substrate specificity assay and stable isotope labeling experiment clearly indicate that AsmC accepts glycine as the substrate, although ashimide requires a serine building block indeed. Interestingly, it appears that asmC and asmD are located in the same operon as the start codon of asmD overlaps with the stop codon of asmC (Scheme 1). AsmD is predicted to be a serine hydroxymethyltransferase that is a pyridoxal-5′-phosphate (PLP) dependent enzyme catalyzing the conversion of glycine to serine by transferring a hydroxymethyl group from 5,10-methylenetetrahydrofolate (mTHF) in primary metabolism.27,28 The ΔasmD mutant abolished the production of 1 and 2 but only accumulated 10 (Fig. 1 and S8†), confirming the necessity of AsmD in ashimide biosynthesis. The co-translation of the genes in the same operon would yield equal amounts of enzymes for their interactions.29 Thus, AsmD could recognize AsmC through a protein–protein interaction and converts AsmC bound glycine to serine. To examine the role of AsmD, we obtained the recombinant AsmD enzyme from E. coli (Fig. S10†) and chemically synthesized N-acetylcysteamine (SNAC) thioesters with l-serine (15) and glycine (16) as mimics of the nascent PCP-tethered amino acids. As mTHF is not commercially available, we carried out the AsmD-catalyzed reverse reaction.28 AsmD was incubated with 15 in the presence of PLP and tetrahydrofolate (THF). Gratifyingly, a new peak with the expected molecular weight of 16 was observed (Fig. 4); in contrast, no reactions occurred when using boiled AsmD as the catalyst. These findings clearly indicate that AsmC activates glycine and recruits AsmD for hydroxymethylation to yield the required serine building block.
Fig. 4. HPLC analysis of the AsmD-catalyzed biochemical reaction in vitro. (i) Standard of 16, (ii) 15 incubated with AsmD in the presence of PLP and tetrahydrofolate, and (iii) l-serine SNAC (15) incubated with boiled AsmD, PLP and tetrahydrofolate.
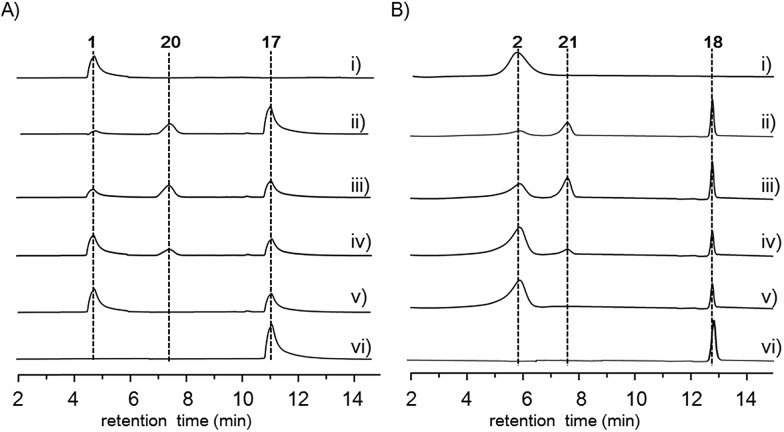
Finally, to investigate the function of the only remaining cytochrome P450 monooxygenase AsmI, we generated the ΔasmI strain. HPLC analysis showed that both final products 1 and 2 were eliminated, leaving 10 and 17–19 as the dominant products (Scheme 1, Fig. 1 and S8, and Tables S17–S19†). Supplementation of 17 and 18 to the ΔasmS mutant was sufficient to restore the production of 1 and 2, respectively, indicating that both compounds are on-pathway biosynthetic intermediates (Fig. S9†). However, 19 is a shunt metabolite or a hydrolyzed artefact as feeding of 19 to ΔasmS could not restore the production of 2 (Fig. S9†). Hence, AsmI is directly involved in the oxidation of the alkene group of benzoxazolinate. However, whether the diol group is directly generated by AsmI or through an epoxide intermediate by spontaneous hydrolysis remains uncertain. To examine this, AsmI was overproduced and purified to homogeneity with an N-terminal his6-tag from E. coli BL21(DE3) (Fig. S10†). The native ferredoxin reductase (FDR) and the ferredoxin (FDX) partner cloned from the same strain S. sp. NA03103 were also overproduced and purified (Fig. S10†). Compound 17 was thus incubated with AsmI in the presence of FDR, FDX and electron donor NADPH for 1 h (Fig. 5A). The production of 1 and a putative epoxide compound 20 with an observed [M + H]+ ion at m/z 447.1145 in LC-MS was observed (Scheme 1, Fig. S13†). Attempts to purify 20 were not successful as it rapidly converted to 1. To further understand this conversion, we carried out a time-course analysis for the AsmI-catalyzed reaction (Fig. 5A). The production of 20 initially increased but was finally converted to 1, suggesting that the epoxide intermediate 20 could be the real natural product, which can be spontaneously hydrolyzed to 1. In addition, AsmI can accept 18 to produce 2 through a putative epoxide intermediate 21 as indicated by its HRESIMS (Scheme 1, Fig. 5B and S14†). Kinetic analysis showed that AsmI displayed similar catalytic activities toward both substrates with Km 2.224 ± 0.2718 mM and kcat 0.166 ± 0.053 min–1 for 17; and Km 2.035 ± 0.2937 mM and kcat 0.171 ± 0.058 min–1 for 18 (Fig. S15 and S16†). In addition, AsmI has no effect on 10 (Fig. S17†).
Fig. 5. HPLC analysis of the AsmI-catalyzed reaction. (A) In vitro assays of AsmI using 17 as the substrate. (i) Standard of 1; 17 was incubated with AsmI, FDR, FDX, and NADPH for (ii) 1 h; (iii) 1.5 h; (iv) 2 h; (v) 3 h; (vi) 17 was incubated with boiled AsmI, FDR, FDX and NADPH for 2 h. (B) In vitro assays of AsmI using 18 as the substrate. (i) Standard of 2; 18 was incubated with AsmI, FDR, FDX, and NADPH for (ii) 1 h; (iii) 1.5 h; (iv) 2 h; (v) 3 h; (vi) 18 was incubated with boiled AsmI, FDR, FDX and NADPH for 2 h.
Taken together, our data revealed that four monomodule NRPSs successively incorporate benzoxazolinate, glycine, glycine and alanine into the assembly line. After conversion of PCP-tethered glycine to serine at the second module by AsmD, the tetrapeptide framework is assembled based on the canonical NRPS rule, followed by N-methylation catalyzed by AsmP. The cytochrome P450 AsmK is expected to desaturate the C21–C25 position to afford 2-aminoacrylic moiety intermediate II,30 which further undergoes hydroxylation/halogenation (both via Fl-4a-OOH as the electrophilic donor) to afford iminium ion intermediates III or III′ (Scheme 1). The transient iminium ion can then be intramolecularly captured by the amine –NH to yield imidazolidin-4-one ring.31,32 The TE domain catalyzes the offloading of the tetrapeptide from AsmN by forming an amide bond to produce 17 and 18, which were finally epoxidized by AsmI to afford the expected final natural products 20 and 21 (Scheme 1). However, both 20 and 21 are prone to be spontaneously hydrolyzed to the isolatable 1 and 2, respectively.
Conclusions
In conclusion, in spite of the great biosynthetic potential of natural products revealed by recent advances in microbial genomics, dereplication and activation gene clusters remain a key issue in obtaining novel secondary metabolites. Our work presented here underpins the importance of combining bioinformatics analysis and heterologous expression of the orphan and neglected gene cluster for novel natural product discovery. The biosynthetic pathway for ashimides A and B isolated in this work was established through gene inactivation, biochemical analysis and stable isotope-labelling experiments, revealing a new desaturation, halogenation/hydroxylation and cyclization cascade to generate structural complexity.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
Authors thank Dr Brian Traw (Nanjing University) for critical reading and helpful discussion. This work was financially supported by the National Natural Science Foundation of China (No. 81522042, 21572100, 81773591, 81530089, 21672101, 81673333, 21861142005, 21761142001, 21661140001, J1210026, and J1103512), Ministry of Science and Technology (2018YFC1706205) and Fundamental Research Funds for the Central Universities (no. 020814380092).
Footnotes
†Electronic supplementary information (ESI) available. CCDC 1861590. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8sc05670f
References
- Marahiel M. A. Nat. Prod. Rep. 2016;33:136–140. doi: 10.1039/c5np00082c. [DOI] [PubMed] [Google Scholar]
- Walsh C. T. Nat. Prod. Rep. 2016;33:127–135. doi: 10.1039/c5np00035a. [DOI] [PubMed] [Google Scholar]
- Bloudoff K., Fage C. D., Marahiel M. A., Schmeing T. M. Proc. Natl. Acad. Sci. U. S. A. 2017;114:95–100. doi: 10.1073/pnas.1614191114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F., Marahiel M. A. Nat. Prod. Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- Süssmuth R. D., Mainz A. Angew. Chem., Int. Ed. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- Strieker M., Tanovic A., Marahiel M. A. Curr. Opin. Struct. Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Walsh C. T. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- Tietz J. I., Schwalen C. J., Patel P. S., Maxson T., Blair P. M., Tai H. C., Zakai U. I., Mitchell D. A. Nat. Chem. Biol. 2017;13:470–478. doi: 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang T. T., Xu Q. L., Xiong Y., Zhang L., Han H., Xu K., Guo W. J., Xu Q., Tan R. X., Ge H. M. Angew. Chem., Int. Ed. 2018;57:8184–8188. doi: 10.1002/anie.201804317. [DOI] [PubMed] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., Lee S. Y., Fischbach M. A., Müller R., Wohlleben W., Breitling R., Takano E., Medema M. H. Nucleic Acids Res. 2015;43:237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M. A., Walsh C. T. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Huang S., Zhao Y., Qin Z., Wang X., Onega M., Chen L., He J., Yu Y., Deng H. Process Biochem. 2011;46:811–816. [Google Scholar]
- Yu L., Mah S., Otani T., Dedon P. J. Am. Chem. Soc. 1995;117:8877–8878. [Google Scholar]
- Liu W., Christenson S. D., Standage S., Shen B. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- Kumada Y., Miwa T., Naoi N., Watanabe K., Naganawa H., Takita T., Umezawa H., Nakamura H., Iitaka Y. J. Antibiot. 1983;36:200–202. doi: 10.7164/antibiotics.36.200. [DOI] [PubMed] [Google Scholar]
- Lalioti M. D., Heath J. K. Nucleic Acids Res. 2001;29:14–19. doi: 10.1093/nar/29.3.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. A., Mavrodi D. V., Thomashow L. S., Roessle M., Blankenfeldt W. J. Biol. Chem. 2011;286:18213–18221. doi: 10.1074/jbc.M110.183418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S. G., Lin S., Shen B. Proc. Natl. Acad. Sci. U. S. A. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang B., Li H., Xie Y., li Q., Qin X., Zhang X., Ju J. Org. Lett. 2015;17:1509–1512. doi: 10.1021/acs.orglett.5b00389. [DOI] [PubMed] [Google Scholar]
- de Mattos-Shipley K. M. J., Greco C., Heard D. M., Hough G., Mulholland N. P., Vincent J. L., Micklefield J., Simpson T. J., Willis C. L., Cox R. J., Bailey A. M. Chem. Sci. 2018;9:4109–4117. doi: 10.1039/c8sc00717a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihlmaier C., Welle E., Hofmann C., Welzel K., Vente A., Breitling E., Mülller M., Glaser S., Bechthold A. Antimicrob. Agents Chemother. 2006;50:2113–2121. doi: 10.1128/AAC.00007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T., Wencewicz T. A. Nat. Prod. Rep. 2013;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. W., Ma M., Kwong T., Ju J., Lim S. K., Jiang H., Lohman J. R., Yang C., Cleveland J., Zazopoulos E., Farnet C. M., Shen B. Biochemistry. 2014;53:7854–7865. doi: 10.1021/bi501396v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttig M., Medema M. H., Blin K., Weber T., Rausch C., Kohlbacher O. Nucleic Acids Res. 2011;39:362–367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottur J., Nair D. T. Nucleic Acids Res. 2018;46:5875–5885. doi: 10.1093/nar/gky402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large P. J., Peel D., Quayle J. R. Biochem. J. 1962;82:483–488. doi: 10.1042/bj0820483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivoli M., Angelucci F., Ilari A., Morea V., Angelaccio S., di Salvo M. L., Contestabile R. Biochemistry. 2009;48:12034–12046. doi: 10.1021/bi901568b. [DOI] [PubMed] [Google Scholar]
- Muliandi A., Katsuyama Y., Sone K., Izumikawa M., Moriya T., Hashimoto J., Kozone I., Takagi M., Shin-ya K., Ohnishi Y. Chem. Biol. 2014;21:923–934. doi: 10.1016/j.chembiol.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Alt S., Wilkinson B. ACS Chem. Biol. 2015;10:2468–2479. doi: 10.1021/acschembio.5b00525. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Haynes S. W., Ames B. D., Gao X., Tang Y. ACS Chem. Biol. 2013;8:1366–1382. doi: 10.1021/cb4001684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. W., Gao X., Tang Y., Walsh C. T. J. Am. Chem. Soc. 2012;134:17444–17447. doi: 10.1021/ja308371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. W., Gao X., Tang Y., Walsh C. T. ACS Chem. Biol. 2013;8:741–748. doi: 10.1021/cb3006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.