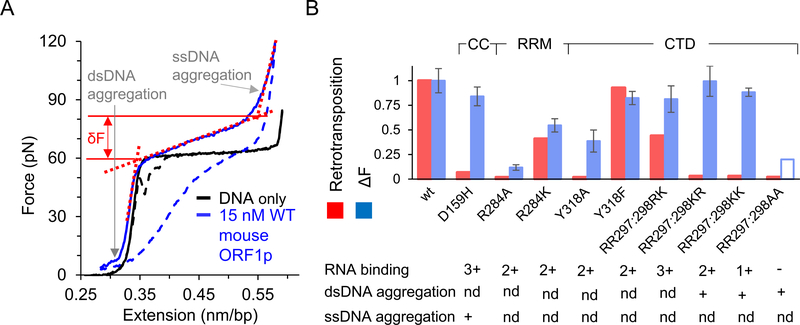
Fig. 5. Single molecule DNA stretching results from mouse ORF1p mutational analyses [41,42,59].
Solid and dashed blue lines are the stretch and return curves of dsDNA in the presence of 15 nM wild type mouse ORF1p. ΔF is the relative increase in the helix-coil transition width due to bound protein, where ΔF = δF–δF0. Here, δF0 (~4pN) and δF are the helix-coil transition widths in the absence and presence of protein, respectively. δF (shown as the force difference between the solid red lines) is determined by the intersection of the dashed red lines on the figure, which represent ds- and ss- DNA regimes of the ORF1p-DNA complex. Thus, ΔF is a measure of the relative increase in force (due to bound protein) required in the cooperative conversion of ds- to ss- DNA, which was shown to be positively correlated with the nucleic acid chaperone capabilities of the bound protein as described in the text. B) ΔF at 15 or 20 nM protein concentration, as measured in single molecule DNA stretching assays (blue), and retrotransposition activity in cultured in vivo assays (red) for ORF1p mutants, presented after normalizing with the corresponding values for the wild type protein. The transition width for the RR297:298AA mutant at these concentrations was not reported due to its lower binding affinity. However, it was shown to saturate at a 5-fold higher concentration than what was used for wild type protein. The empty bar is to denote that the transition width is likely much smaller at these concentrations in comparison with the wild type protein. Extensive aggregation of single- or double- stranded DNA in comparison with the wildtype protein, are denoted with a plus sign. While aggregation is important to facilitate DNA interactions and can increase chaperone activity, extensive aggregate formation can also slow down DNA interaction kinetics, which may in turn inhibit chaperone activity [79]. The ability to bind RNA as observed in the bulk solution assays is also reported. The plus sign denotes comparable binding affinity as wild type and these mutants are ranked according to their relative binding affinities, with (−) the lowest and (3+) the highest affinity. ‘nd’ represents ‘not determined’.