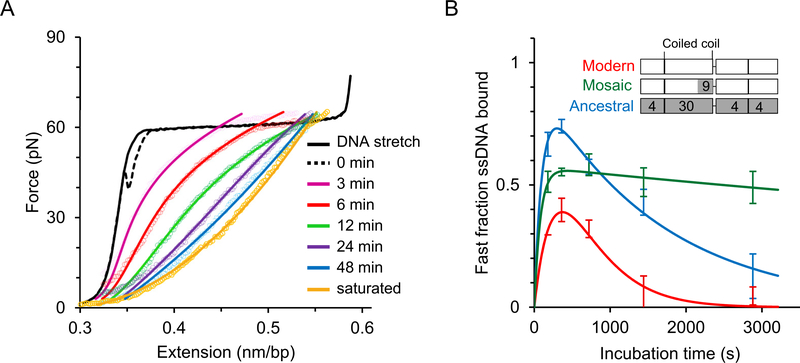
Fig. 6. Single molecule studies of human ORF1p [43].
A) In contrast to the studies described in Fig. 5, here, the DNA molecule is overstretched above the helix-coil transition up to the ssDNA regime before incubating it with the protein, in order to minimize dsDNA binding effects and exclusively investigate ssDNA-ORF1p binding kinetics. Solid and dashed line represent the stretch-return cycle of a bare dsDNA, with the return curve also referred to as 0 min incubation. The colored empty circles are the return curves of a DNA molecule after incubating with 2 nM protein at ~70 pN for different time periods. The gold curve listed as saturated represents the maximum shift in extension towards ssDNA, obtained at long incubation times and greater than 15 nM protein concentration. The ssDNA-bound protein during the incubation prevents reannealing and thereby shift the relaxation curve after protein incubation towards the ssDNA curve (see green circles, Fig. 4B), allowing one to quantitatively probe the ssDNA fraction bound as a function of time. The solid lines represent the fitted curves as described elsewhere [43]. ORF1p rapidly binds ssDNA. However, it transforms relatively slowly into stable oligomers. Less stable proteins (presumably un-transformed trimers) dissociate during the return cycle and the ssDNA fraction bound by such proteins is defined as the fast fraction in this study. The fast fraction decreases with incubation time as they transform into more stable oligomers on ssDNA. B) Fast fraction as a function of time for modern human (111p, from L1Pa1 family), its resuscitated ancestral primate (555p, from L1Pa5 family) and a mosaic ORF1p (151p) in which 9 residues in the coiled coil are replaced with the corresponding ancestral residues, as shown in the inset (also see Fig. 2 where these residues are indicated in teal). Domain boundaries in the inset correspond to Fig. 1B and white and grey shades represent the amino acid residues of the modern and ancestral ORF1p, respectively. The number of amino acid substitutions relative to the modern ORF1p is denoted in the relevant domains in the other two ORF1p variants. The measured fast fraction is modeled as a sum of increasing and decreasing exponential functions (solid lines) and red, blue and green represent modern, ancestral, and mosaic proteins, respectively. The fast fraction rapidly saturates for all three variants, indicating rapid protein binding to ssDNA. However, this fraction decreases with increasing incubation time as the proteins form more stable oligomers. Therefore, the rate at which the fast fraction decreases is proportional to the rate of stable oligomerization of protein on ssDNA. The retrotransposition-incompetent mosaic ORF1p was at least 10-fold slower in forming stable oligomers in comparison with the active human and primate protein variants.