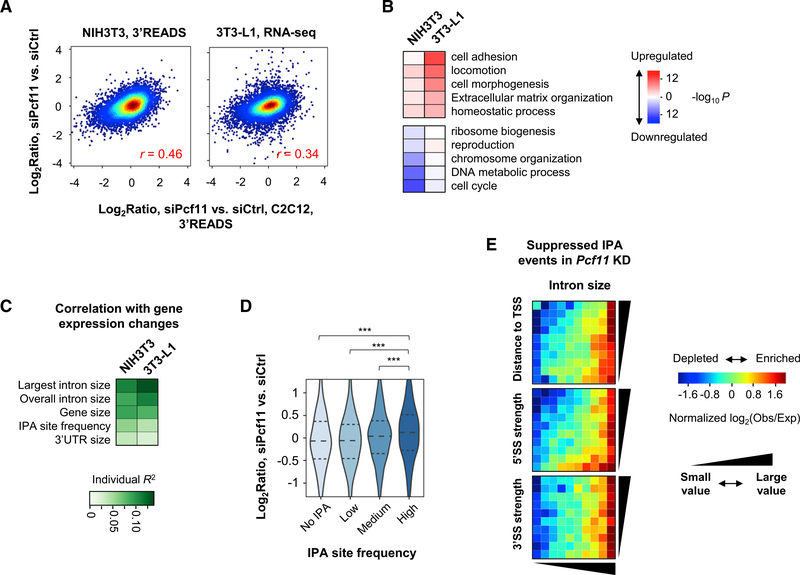
Figure 5. Gene Expression Regulation by PCF11 in Different Cell Contexts.
(A) Correlation of gene expression changes by Pcf11 KD in C2C12 cells versus NIH 3T3 (left) or 3T3-L1 (right) cells. NIH 3T3 data are based on 3′READS and 3T3-L1 data on RNA-seq.
(B) Top GO terms associated with up- or downregulated genes in NIH 3T3 and 3T3-L1 Pcf11 KD cells.
(C) Top gene features correlated with gene expression changes by Pcf11 KD in NIH 3T3 and 3T3L1 cells.
(D) mRNA expression changes in NIH 3T3 KD cells for genes with different IPA site frequencies (number of IPA sites per gene) as annotated in PolyA_DB. ***p < 0.001 (Wilcoxon test).
(E) IPA distribution maps for suppressed IPA events in NIH 3T3 KD cells.