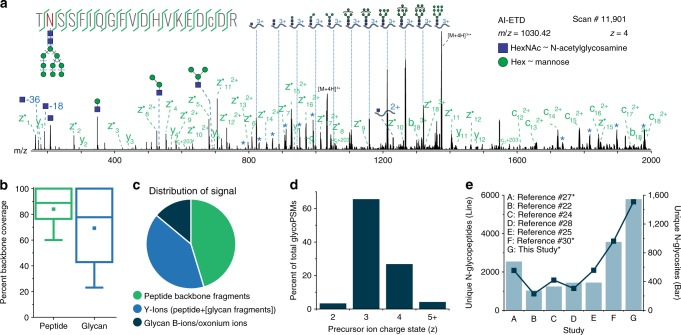
Fig. 1.
Identifying intact glycopeptides with AI-ETD. a Annotated single AI-ETD spectrum (i.e., no averaging) of N-glycopeptide TN*SSFIQGFVDHVKEDcDR modified with a high mannose-type glycan [HexNAc(2)Hex(9)]. The red asparagine indicates the site of glycosylation, and the lowercase cysteine indicates carbamidomethylation. Green fragments are products from peptide backbone cleavage, triply charged Y-ions are annotated along the top, and B-ions include only glycan moieties. Blue asterisks (*) denote doubly and quadruply charged Y-ions (from 1700 to 2000 and 750 to 1000 Th, respectively), each which differ by one hexose residue. Peptide fragments retain the glycan modification unless denoted by a “~”. b Distribution of percent peptide backbone coverage and glycan coverage seen in AI-ETD spectra. Median and quartile values are provided by the center line and box boundaries, respectively. Whiskers show 10th and 90th percentiles, and the small square indicates the average. c Average percent of explained ion current in product ions in AI-ETD spectra from peptide backbone cleavage fragments, Y-ions (i.e., intact peptide sequence with fragments of the glycan moiety), and B-ions/oxonium ions. d Distribution of precursor ion charge states successfully identified in the 24,099 glyco PSMs from this study, given as a percentage of the total. e Comparison of recent large-scale N-glycopeptide studies showing the number of unique N-glycopeptides (left axis, dark blue line) and unique N-glycosites (right axis, light blue bars) identified. Asterisks (*) by the study name indicate that mouse brain was the system investigated