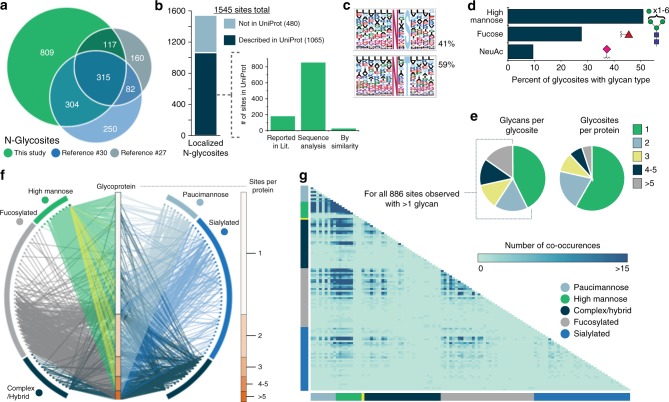
Fig. 2.
Characteristics of glycosites identified with AI-ETD. a Overlap of mouse brain N-glycosites identified in this study with those from Liu et al.30 and Trinidad et al.27 studies. b Approximately 69% of identified glycosites are described as known glycosites in the UniProt database, and the majority of them have that description based on sequence analysis (i.e., prediction of glycosite based on the presence of the N-X-S/T sequon). c Sequence motifs for N-glycosites having either the N-X-S or N-X-T sequon and their relative percentage in the unique glycosites identified. d Percentage of total glycosites that had glycans of high mannose type or that contained a fucose or NeuAc residue. e Distribution of the number of different glycans seen at a given glycosite, i.e., the degree of glycan microheterogeneity (left), and the number of glycosites per glycoprotein identified (right). f A glycoprotein-glycan network maps which glycans (outer circle, 117 total) modify which proteins (inner bar, 771 total). Glycoproteins are sorted by number of glycosites (scale to the right). Glycans are organized by classification, and edges are colored by the glycan node from which they originate, except for mannose-6-phosphate which has yellow edges. See Supplementary Fig. 11 and Supplementary Table 1 for glycan identifiers. g A glycan co-occurrence heat map represents the number of times glycan pairs appeared together at the same glycosite, indicating which glycans contribute most to microheterogeneity of the >880 sites that had more than one glycan modifying them