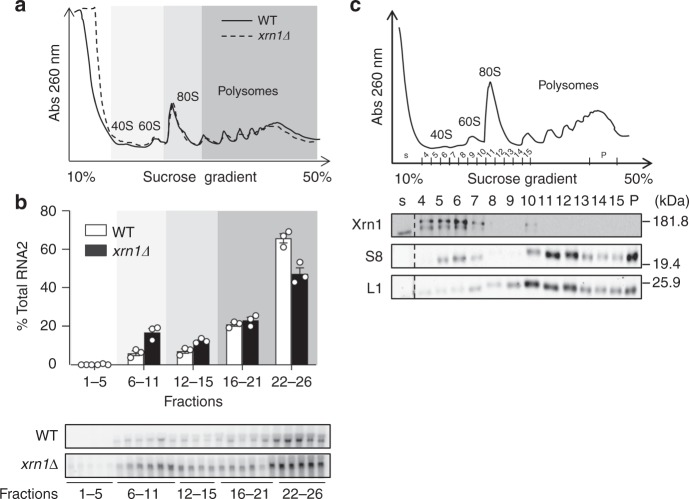
Fig. 2.
Xrn1 depletion shifts RNA2 toward single ribosomal subunits fractions. a ultraviolet (UV) absorbance rRNA profile at 260 nm of an extract from WT and xrn1∆ cells expressing RNA2 after sedimentation on a 10 to 50% sucrose gradient. b Depletion of Xrn1 shifts BMV RNA2 toward monosomal fractions. Upper panel: distribution of normalized BMV RNA2 accumulation across the profiles. Fractions were grouped into free (1–5), single ribosome subunits (6–11), monosomes (12–15), light polysomes (16–21), and heavy polysomes (22–26). RNA was quantified by northern blot. Results represent averages of n = 3 biological replicates. Error bars represent SEM. Open circles indicate the individual data points. Lower panel: representative northern blots. c Xrn1 cofractionates with free 40s subunits in polysome-profiling analysis. Top panel: UV absorbance rRNA profile at 260 nm of an extract from WT cells. Lower panel: Fractions were TCA-precipitated and analyzed by western blot. Specific antibodies detecting Xrn1p, S8 protein (small ribosomal subunit) and L1 protein (large ribosomal subunit) were used. S lane corresponds to soluble proteins not associated to ribosome subunits and P lane to a pool of three fractions corresponding to polysomes. Source data are provided as a Source Data file