Abstract
Atopic dermatitis (AD) has been hypothesised to be associated with gut microbiota (GM) composition. We performed a comparative study of the GM profile of 19 AD children and 18 healthy individuals aimed at identifying bacterial biomarkers associated with the disease. The effect of probiotic intake (Bifidobacterium breve plus Lactobacillus salivarius) on the modulation of GM and the probiotic persistence in the GM were also evaluated. Faecal samples were analysed by real-time PCR and 16S rRNA targeted metagenomics. Although the probiotics, chosen for this study, did not shape the entire GM profile, we observed the ability of these species to pass through the gastrointestinal tract and to persist (only B. breve) in the GM. Moreover, the GM of patients compared to CTRLs showed a dysbiotic status characterised by an increase of Faecalibacterium, Oscillospira, Bacteroides, Parabacteroides and Sutterella and a reduction of short-chain fatty acid (SCFA)-producing bacteria (i.e., Bifidobacterium, Blautia, Coprococcus, Eubacterium and Propionibacterium). Taken togheter these results show an alteration in AD microbiota composition with the depletion or absence of some species, opening the way to future probiotic intervention studies.
Introduction
In the last decades, an increase in allergic diseases has been observed worldwide, especially in westernised countries1. Among allergies, atopic dermatitis (AD) is the most common chronic inflammatory skin disease that occurs early in life with a prevalence of 20% in children. The disease has a deep impact on the quality of life of the patient and family2. According to the hygiene hypothesis, the effects of modern public health practices, which lower the stimulation of the immune system by microbes, make infants more likely to develop allergic diseases3,4. As an extension of the hygiene hypothesis, the “microflora hypothesis of allergic disease” was proposed to underline the role of the gut microbiota (GM) in shaping the development of the host immune system in early life5. In fact, the early exposure to gut microbes shifts the Th1/Th2 balance to a Th1 phenotype6,7. In contrast, the absence of a normal intestinal bacterial colonisation in atopic diseases, particularly during the mucosal immune system development, pushes the Th1/Th2 balance towards a Th2 response8,9. Th2 cell-derived mediators, such as IL-4, IL-5 and IL-13, induce immunoglobulin class switching to IgE, thus sustaining the allergy response. Modulation of this response through T-cell deviation to Th2 or enhancement of regulatory T-cells (Treg) is a new therapeutic strategy for the prevention and treatment of AD through probiotic administration10. Probiotics are defined as living microorganisms that, once ingested, exert health benefits for the host11. In the context of atopic diseases, they may act as immune modulators that stimulate Th1-mediated responses12. The positive effects of probiotics on AD symptoms is already described in literature13,14 and, indeed, probiotics administration is routinely employed in clinical practice.
However, recommendations on timing and dose of administration of probiotics in AD have not yet been established. Moreover, not all the studies based on probiotic intake demonstrate the actual ability of probiotics to colonize the GT. This aspect does not allow clinicians to choose the probiotic strain with the certainty that it persists in the GM. In the present study, GM of AD patients and healthy age-matched controls were profiled to identify bacterial biomarkers associated with the disease. The effect of the intake of probiotics composed by Bifidobacterium breve and Lactobacillus salivarius was evaluated with respect to GM modulation over time to assess the persistence of the probiotic bacteria by quantitative Real-Time PCR (qRT-PCR).
Materials and Methods
Study design and sample collection
Nineteen patients in the age range of 0 to 6 years with a diagnosis of AD were prospectively enrolled in the study at the Dermatology Unit of the Bambino Gesù Children’s Hospital in Rome, Italy. Patients took the probiotic (composed of B. breve BR03 and L. salivarius LS01) twice per day (1 × 109 UFC/dose of each species for 20 consecutive days). Exclusion criteria consisted of treatment with steroids or calcineurin inhibitors, antibiotics intake or gastrointestinal disorders in the four weeks before enrolment and during the follow up. Faecal samples were collected during clinical visits at time T0 (no probiotic intake), T1 (completion of probiotic intake), T2 (60 days after the end of probiotic intake); T3 (90 days after the end of probiotic intake).
The treatment was based on antiseptics, emollients, hydration and also focused on therapeutic patient education. In only two patients it was necessary to recurr to topyc steroids during the last two time points.
Seventy-two faecal samples were collected and accompanied by a clinical data diary, SCORAD index values, comorbidities, vaccinations and diet type. The samples were stored at −80 °C at the Human Microbiome Unit of Bambino Gesù Children Hospital in Rome until DNA extraction. Eighteen faecal samples from healthy children of the same age range were also collected as age-matched controls (CTRLs). The criteria for the CTRLs were the absence of chronic diseases or gastrointestinal infections and no antibiotic or probiotic intake in the four weeks before the enrolment. This study was approved by the OPBG Ethics Committee (protocol number 391LB).
All research was performed in accordance with relevant guidelines/regulations. Informed consent was obtained from all parents and/or legal guardians of participants.
Isolation of B. breve BR03 and L. salivarius LS01 bacterial strains from the probiotics
The B. breve BR03 and L. salivarius LS01 bacterial strains were cultured on Columbia agar +5% sheep blood medium (COS, Biomerieux Marcy l’Etoile, France) at 37 °C for 24 h under anaerobic (B. breve) or aerobic (L. salivarius) conditions. Single colonies were isolated and purified on new COS plates based on their morphology. Bacterial identification was confirmed using a matrix assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) biotyper and a Microflex LT mass spectrometer (Bruker Daltonics, Bremen, Germany) as described15.
Bacterial DNA extraction from colony and 16S ribosomal locus amplification
DNA was extracted from bacterial colonies using the EZ1 DNA Tissue Kit and automatic extractor biorobot EZ1 according to the manufacturer’s instructions (Qiagen, Hilden, Germany). The entire 16S rDNA locus (1465 bp) was amplified from the extracted DNA using universal primers (27f 5′-AGAGTTTGATCCTGGCTCAG-3′/1492r 5′-ACGGTTACCTTGTTACGACTT-3′). PCR was performed with a reaction mixture containing 5 µl 10X Buffer, 2 µl 2.5 mM MgCl2, 2 µl each primer (10 µmol/L), 2 µl dNTPs (10 mmol/L), 1 µl Taq DNA polymerase (5 U/µl) (KAPA Taq PCR kit, KAPA Biosystems, Boston, USA), 5 µl DNA template (10 ng/µl) and molecular-grade H2O to a final reaction volume of 50 µl16. The amplification protocol consisted of one cycle of initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min followed by a final extension at 72 °C for 10 min. The resulting amplicons were purified using centrifugal filter units (Amicon Ultra-0.5 mL Centrifugal filters 30 K, Sigma-Aldrich, MO, USA) and quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, DE, USA).
Bacterial DNA cloning and species-specific primer and probe design
The purified PCR products were sequenced and cloned into the PGEM vector following the instructions provided by the pGEM®-T Easy Vector System kit (Promega, Italy) using Escherichia coli competent cells as a host. The obtained plasmids, pGEM-BB (pGEM + B. breve) and pGEM-LS (pGEM + L. salivarius), were extracted (Plasmid Miniprep Kit, Promega, Italy), quantified using the NanoDrop ND-1000 spectrophotometer and diluted. The dilutions, which ranged from 106 to 101 vector copy numbers, were used as standards in the quantitative RT-PCR (qRT-PCR) assays. The cloned fragments of 16S rDNA in the pGEM-BB and pGEM-LS vectors were amplified and sequenced with an automated sequence analyser (Genetic Analyser 3500, Applied Biosystems, CA, USA) using a 50-cm capillary array and a POP-7 polymer (Applied Biosystems) and the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, version 3.1) according to the manufacturer’s instructions. All electropherograms were manually edited for base ambiguity. The obtained FASTA sequences were aligned using CLUSTAL-W software (http://www.ebi.ac.uk/clustalw/) and used for the design of species-specific primers and TaqMan probes (Roche Diagnostics, Mannheim, Germany) (Supplementary Table 1).
Bacterial DNA extraction from stool samples
Frozen stool samples were thawed at room temperature, and DNA was manually extracted using the QIAmp Fast DNA Stool mini kit (Qiagen, Germany) according to the manufacturer’s instructions. DNA was quantified using the NanoDrop ND-1000. Comparable amounts of DNA (80 ng) from each sample were used in the qRT-PCR assays.
Targeted-metagenomics
The V1–V3 regions (520 bp) of the 16S ribosomal RNA (rRNA) locus were amplified and pyrosequenced using a 454-Junior Genome Sequencer (Roche 454 Life Sciences, Branford, USA)17. The obtained raw reads were analysed using Quantitative Insights into Microbial Ecology software (QIIME) 1.8.0 software18 by demultiplexing, quality score checking, low-length excluding and denoising19. Sequences were grouped into operational taxonomic units (OTUs) by clustering at a threshold of 97% pairwise identity using UCLUST for sequence clustering20 and the representative sequences were submitted to PyNAST for sequence alignment18. The Greengenes database (v 13.8) was used for OTU matching.
qRT-PCR
Quantification of B. breve and L. salivarius in faecal samples was carried out by qRT-PCR using the Light Cycler 480 platform (Roche Diagnostics, Mannheim, Germany). The assays were performed with a 20 µl PCR amplification mixture containing: 10 µl LightCycler 480 Probe Master mix (Roche Diagnostics), 2 µl primers and probes (optimized concentrations, 0.5 µM and 0.1 µM, respectively) (Supplementary Table 1), 3 µl molecular-grade H2O and 5 µl DNA template. Each sample was tested in duplicate to ensure data reproducibility. The RT-PCR temperature profile consisted of an initial denaturation at 95 °C for 10 min, 45 amplification cycles at 95 °C for 10 sec, 60 °C for 30 sec and 72 °C for 1 sec followed by a final cooling step at 40 °C for 30 sec. Absolute quantification was performed using the “second derivative maximum method”21.
Statistical analyses
All data were tested for normal distribution using the Shapiro-Wilk normality test. Statistical analyses were computed using the phyloseq R package for alpha and beta diversity22. Furthermore, the adonis function in the vegan R package was used to perform the PERMANOVA test on beta diversity with 999 permutations using the “strata” argument within the adonis function.
The non-parametric Mann-Whitney U-test and Wilcoxon signed-rank test were used to compare the two independent groups (CTRL versus AD at T0) and the data for the time points within the AD group, respectively. The linear discriminant effect size (LEfSe) was computed23 with α value equal to 0.05 and a logarithmic LDA score threshold of 2.0. The area under the relative operating characteristic (AUROC) test and discriminant analysis (DA) based on univariate ANOVA, Fisher’s coefficient and leave-one-out classification were applied. Correlations between B. breve and L. salivarius concentrations were determined by the Spearman test using IBM SPSS statistical software (version 21). Only significant p-values (p < 0.05) corrected using the Holm method were considered24.
Metagenomic data open access repository
All sequences and the associated metadata are available at NCBI: Bioprojects: PRJNA439447, gut metagenomic profile from AD patients; PRJNA268064, gut metagenomic profile from healthy subjects (http://www.ncbi.nlm.nih.gov/bioproject/?term=).
Results
This study included 19 AD patients and 18 CTRLs, age 0 to 6 years. Patient and CTRL characteristics are summarized in Table 1. In particular 12/19 were vaginal delivered, 13/19 were breast-fed and 15/19 were weaned after 6 months.
Table 1.
Demographic characteristics of patients and healthy individuals enrolled in this study.
| AD group | CTRL group | |
|---|---|---|
| N | 19 | 18 |
| Male, N | 12 | 11 |
| Female, N | 7 | 7 |
| Age, years (±sd) | 2.2 (1.7) | 2 (1.4) |
| BMI (±sd) | 16.3 (1.9) | 15.95 (1.4) |
| Vaginal delivery, N | 12 | n.a |
| Caesarean section, N | 7 | n.a |
| Breastfeeding, N | 13 | n.a |
| Formula feeding, N | 6 | n.a |
| weaning time <6 months, N | 4 | n.a |
| weaning time ≥6 months, N | 15 | n.a |
Targeted-metagenomics
Eighty-eight faecal samples were analysed by 16S targeted-metagenomics with two samples excluded due to the poor quality of the metagenomic reads, which consisted of 18 baseline (T0) and 52 follow-up (19 at T1, 17 at T2, 16 at T3) samples from the AD patients and 18 from the CTRL subjects. From the total set of samples, 239,153 sequencing reads with a mean value of 2,202 sequences for each sample were obtained.
The microbiota biodiversity of the AD and CTRL groups was determined using alpha and beta diversity analyses. In the comparison between the AD and CTRL groups, we tested the effects of the clinical variables (i.e., delivery, neonatal feeding before weaning, weaning time, age, and BMI) on the AD sample distribution. Beta diversity analysis (Bray-Curtis, unweighted and weighted UniFrac metrics) which included all these variables, showed that “age” and “weaning time” greatly influenced the distribution of samples in the principal coordinate analysis (PCoA). Patients under the age of one year (infants) differed enormously from the rest of the sample cohort and formed a significantly separated cluster (Supplementary Fig. 1, Supplementary Table 2). To avoid bias in the comparison between the AD and CTRL groups, we excluded patients under the age of 1 year, thus eliminating the age and weaning time effects (Supplementary Table 3). Hence, final analysis included 15 AD (patients older than 1 year of age) and 18 CTRLs.
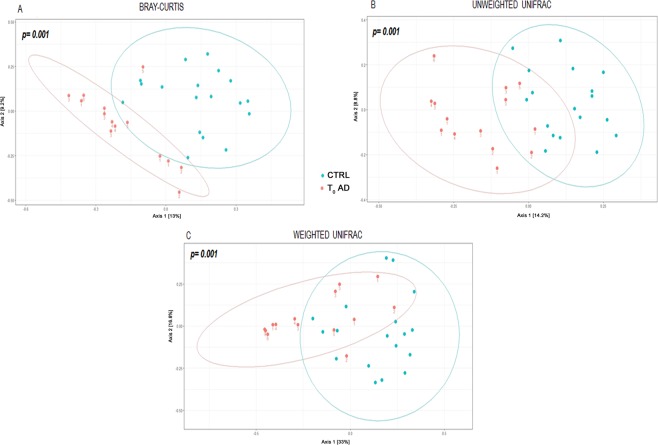
To evaluate how OTUs were differentially distributed in the AD and CTRL groups, differences in beta diversity were calculated. A clear separation was observed between the two groups (Fig. 1), which was verified using the PERMANOVA test (p = 0.001 for weighted, unweighted and Bray-Curtis analyses). However, there were no clearly defined clusters for the patients stratified into T0 and T1–T3 groups (PERMANOVA > 0.05) (Supplementary Fig. 2).
Figure 1.
Beta-diversity analysis of AD and CTRL groups. The plots show the first two principal axes for PCoA using Bray-Curtis (A), unweighted UniFrac (B) and weighted UniFrac (C) algorithms. P-values were obtained by PERMANOVA.
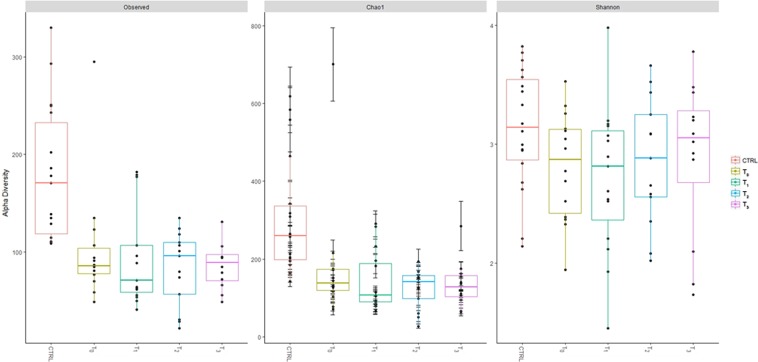
Alpha diversity was calculated with respect to OTU richness, evenness and rarity to understand the ecological differences within the AD and CTRL groups using the Shannon, observed and Chao1 indices, respectively. AD patients showed a significantly lower level of alpha biodiversity according to the observed and ChaoI indices compared to the CTRLs at each time point (T0, T1, T2, T3) (Fig. 2, Supplementary Table 3). In addition, the Shannon index revealed less biodiversity in the AD patients, but this result did not reach statistical significance. No statistically significant differences amongst T0–T3 time point paired comparison were achieved (Supplementary Table 3).
Figure 2.
Alpha-diversity measures of Observed, Chao1 and Shannon indexes. Boxes represent the median, 25th and 75th percentiles for AD (time points are indicated) and CTRL groups.
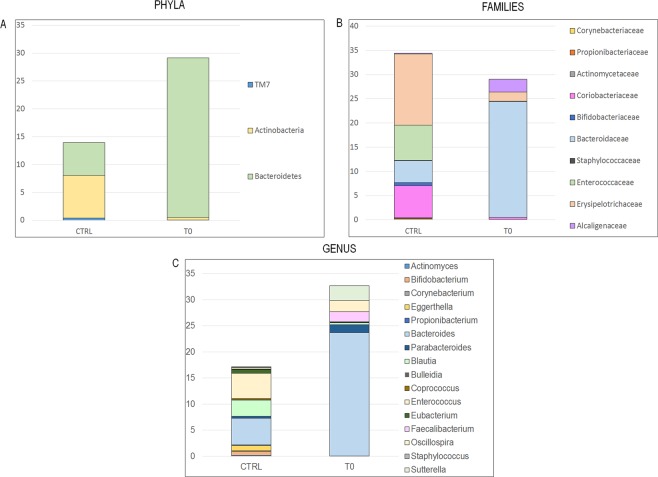
To detect differences in OTU composition between AD patients and the CTRLs, we compared time point T0 of the AD group (i.e., before the probiotic intake) to the CTRL group. The OTU distribution was investigated at the phyla, family and genus levels. At the phylum level, the Mann-Whitney U-test highlighted the OTU abundance differences, which showed that Bacteroidetes was significantly higher in the T0 AD group and Actinobacteria and TM7 were significantly increased in the CTRLs (Fig. 3, Panel A; Supplementary Table 4).
Figure 3.
Mann–Whitney-based OTU distribution. The bar graphs represent the average distribution of the OTUs by phylum (A), family (B) and genus/species (C) levels. Only statistically significant OTUs are plotted (p-values were corrected by the Holm method).
At the family level, the AD group was characterised by lower relative abundances of the Actinobacteria families, such as Propionibacteriaceae, Actinomycetaceae, Coriobacteriaceae and Bifidobacteriaceae, and the complete absence of Corynebacteriaceae (Fig. 3, Panel B; Supplementary Table 5). Moreover, the Firmicutes families also showed different distributions between the two groups. In particular, Erysipelotrichaceae and Enterococcaceae were substantially reduced in AD patients, and Staphylococcaceae was completely absent. In contrast, these patients were highly enriched in Bacteroidaceae (Bacteroidetes) compared to the CTRLs, which accounted for up to 23% of the overall families followed by the Proteobacteria family Alcaligenaceae (2.6%) (Fig. 3, Panel B; Supplementary Table 5).
At the genus level, the AD group showed higher relative abundances of Sutterella (Alcaligenaceae), Bacteroides (Bacteroidaceae), Parabacteroides (Porphyromonadaceae), Oscillospira and Faecalibacterium (Ruminococcaceae, Clostridia Class), and lower relative abundances of Eggerthella (Coriobacteriaceae), Propionibacterium (Propionibacteriaceae), Enterococcus (Enterococcaceae), Eubacterium (Erysipelotrichaceae), Actinomyces (Actinomycetaceae), Blautia and Coprococcus (Lachnospiraceae). Some OTUs, such as Staphylococcus (Staphylococcaceae), Bifidobacterium (Bifidobacteriaceae) Corynebacterium (Corynebacteriaceae) and Bulleidia (Erysipelotrichaceae) were completely absent (Fig. 3, Panel C; Supplementary Table 6). To validate our results, we applied LEfSe on our OTU table in the comparison between AD T0 and CTRL. Mann-Whitney U-test and LEfSe analyses showed consistent results (Supplementary Fig. 3).
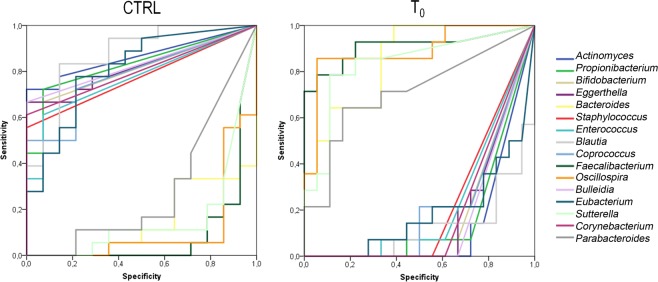
To understand the 16 OTUs belonging group, we applied the average area under the relative operating characteristic (AUROC) test. According to the classification proposed by Swets (Swets 1988), AUROC values ranging from 0.7 to 1 were considered accurate to discriminate between the groups. This analysis showed that the 16 OTUs had discriminatory power. In particular, Bacteroides, Parabacteroides, Faecalibacterium, Oscillospira and Sutterella were selectively associated with the AD T0 group, and Actinomyces, Propionibacterium, Bifidobacterium, Eggerthella, Staphylococcus, Enterococcus, Blautia, Coprococcus, Bulleidia, Corynebacterium and Eubacterium were discriminatory for the CTRL group (Fig. 4, Supplementary Table 7).
Figure 4.
ROC curve plots. The areas under the ROC curves (AUROC) represent the specificity and sensitivity of the 17 selected OTUs to discriminate the T0 AD and CTRL groups.
To estimate the statistical power of these OTUs to act as a classifier for each group, we used the DA model. The DA revealed that 96.9% of the original groups and 84.4% of the cross-validated groups were correctly classified (Supplementary Table 8).
To evaluate the influence of probiotic intake on gut microbiota modulation, we tested the OTU distribution using the pairwise Wilcoxon signed-rank test to compare all time point samples. The test did not find any significant differences at any taxonomic level.
RT-PCR analysis
RT-PCR was performed on AD patient samples (15 T0, 15 T1, 14 T2 and 11 T3 from >1-year-old patients) using primers and probes specific for B. breve and L. salivarius. The median concentrations (molecules/ul) of the two species were higher at T1 than the other time points (Fig. 5). B. breve persisted until the latest time point (T3) whereas L. salivarius decreased to zero by time point T2 (Fig. 5). In particular, the B. breve concentration differences were statistically significant for the pairwise comparisons between T0 and all of the follow-up time points. In contrast, L. salivarius concentration achieved significant differences between T1 and all the other time points (Supplementary Table 9).
Figure 5.
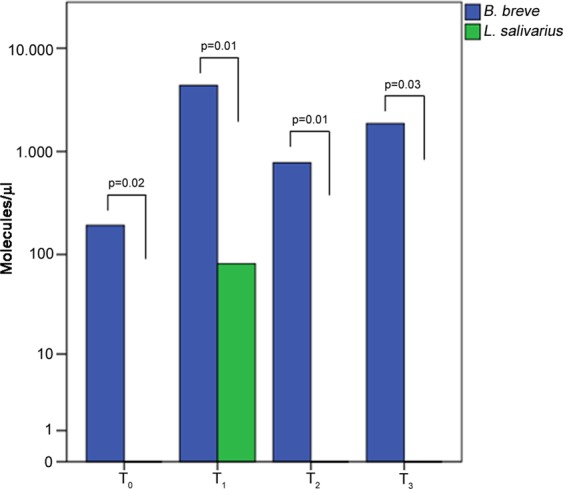
Histograms of B. breve and L. salivarius levels at different time points. Median values differences between B. breve (blue bar) and L. salivarius (green bar) levels expressed as molecules/ul at each point of the time-course. P-values were obtained using the Mann–Whitney U test.
Correlation analysis
A Spearman’s rank correlation analysis was carried out for each time point to understand the relationship between the B. breve and L. salivarius levels. Interestingly, at the T1 time point, we observed a significant positive correlation between B. breve and L. salivarius, suggesting that the species cooperated in their tendency to increase (Supplementary Table 10). No other statistically significant correlations were found.
Discussion
GM dysbiosis has been shown to precede the onset of AD25. However, studies on atopic diseases and microbiota are conflicting because both decreased and increased bacterial community diversity have been related to atopy26. Our results were consistent with previous findings of low intestinal microbial diversity in AD27–29, which supports the theory of ‘microbial deprivation syndromes of affluence30. According to this theory, reduced intensity and diversity of microbial stimulation lead to an abnormal immune maturation in early childhood. In fact, limited microbial pressure results in insufficient Th1 cell induction and the failure to suppress Th2 responses. The switching of the immune stimulation towards a pronounced Th2-phenotype is suggested to be a major mechanism to explain allergy development and maintenance30,31.
Studies focused on the intestinal microbiota composition in AD children are also contradictory32–35.
In our study, we identified some OTUs associated with AD, such as Faecalibacterium and Oscillospira (Firmicutes), Bacteroides and Parabacteroides (Bacteroidetes) and Sutterella (Proteobacteria).
Faecalibacterium genus is generally an indicator of the healthy status of the gut due to its anti-inflammatory effects36,37. Low levels of Faecalibacterium and in particular of F. prausnitzii in the GM have been associated with Crohn’s disease38 and AD children33. A possible explanation of the high levels of this genus in AD, reported by Song and co-workers37, is that the inflamed epithelium with a barrier dysfunction, which is typical in AD, can release nutrients that stimulate the growth of F. prausnitzii subspecies that are not short-chain fatty acid (SCFA)-producing. The decreased production of SCFAs, such as butyrate, could lead to further inflammation in the gut epithelium.
Oscillospira is a common inhabitant of the GM. It is a butyrate-producing bacterium that utilizes host glycans as growth substrates and contributes to the maintenance of gut health39. The role of Oscillospira in atopy is not clear. However, our results show that it is strongly associated with AD, which is consistent with the results of Canani and co-workers32. Its presence in the GM of AD children could likely be linked to the high abundance of Bacteroidaceae, which produce fermented products that serve as substrates for Oscillospira growth40.
Bacteroides spp. are common inhabitants of the human gut, however their increased presence has been associated with food allergy and other atopic manifestations41–43. Indeed, higher levels of Bacteroides in atopy could lead to the continuous production of lipopolysaccharides (LPS), the major component of gram-negative cell wall, in the gut, which could trigger an inflammatory response44. Moreover, Bacteroides species are reported to alter gut permeability41,45, a condition observed in AD.
Among the Proteobacteria, Sutterella levels were increased in AD children. This genus has been associated with other inflammatory diseases, such as Crohn’s disease and ulcerative colitis, but also with healthy adults46,47. Thus, it is still not completely clear if Sutterella is involved in inflammation or is a normal inhabitant of the human GM.
Our results revealed that AD GM is characterised by low colonisation of OTUs that have a role in the maintenance of gut health, like Actinomyces and Eggerthella.
Actinomyces spp are members of the normal oral microbiota and have been reported as one of the causative bacteria of dental caries and periodontal diseases48. Several studies reported that Actinomyces spp. are early colonisers of the healthy infant gastrointestinal tract (GT)49.
Eggerthella belongs to the Coriobacteria that are common members of the human GM50. They are assacharolytic bacteria that produce formate and lactate only from glucose49. Until now, only Eggerthella lenta and the still not fully characterized Eggerthella spp. YY7918 are associated with the human gastrointestinal tract49. Morinaga (1988) studied the role of E. lenta in the stimulation of the immune system and found that this bacterium is involved in the production of anti-tumour molecules that stimulate natural killer cells51. Furthermore, E. lenta could have a role in the stimulation of hepatic detoxification and in the inactivation of pharmaceuticals in the gut52.
Moreover, AD patients showed a strong reduction of some SCFA-producing bacteria, such as Bifidobacterium, Blautia, Coprococcus, Eubacterium and Propionibacterium53,54. SCFAs are of particular interest for maintaining host health because they may exert anti-inflammatory effects through several mechanisms, including epithelial integrity (preserving tight junctions) and maintenance of the mucus layer55. The resulting low production of SCFAs may be causative of the intestinal barrier dysfunction, increased intestinal permeability and inflammation found in ADs56. In particular, Coprococcus spp. are butyrate-producing bacteria49. Interestingly, Nylund and co-workers reported an inverse correlation between the SCORAD index and the levels of Coprococcus eutactus, confirming the role of this particular genus in the amelioration of AD29.
Bifidobacterium spp. are assumed to be beneficial for human health due to their several effects such as vitamin production, immune system stimulation, inhibition of potentially pathogen bacteria, improvement of food ingredients digestion57,58. In the contest of allergic diseases, several studies based on murin and in vitro models, have higlighted the potential role of Bifidobacterium in reducing inflammation by inducing the production of anti-inflammatory cytokines and suppressing Th2 immune response and IgE production25,59,60. The absence of Bifidobacterium in AD children is consistent with other studies35,61 and could lead to a lack of anti-inflammatory effects.
Existing treatments for AD are limited. Therefore, the focus is now to identify alternative strategies. Based on the hygiene hypothesis, probiotics have been proposed as therapeutic and preventive interventions for allergic diseases with the aim to attenuate inflammation62. The most used species, belonging to Bifidobacteria and Lactobacilli, have been shown to reduce the risk of AD63.
Because of controversial results64,65, the influence of probiotics on the prevention or management of AD requires further investigation. In particular, the recommendations on the time of administration and the dose of the probiotic in AD have not been yet drawn up. Huang and collegues13 reported a metanalysis of the data available on the topic, highlighting a multitude of administration approaches, including different doses and intake timing. For this reason we investigated the persistence of the two probiotic strains in the GT at defined time points. In our study, we observed an increase of B. breve and L. salivarius in the stool samples starting from time point T1. The significant increase of these species at the end of the probiotic intake, indicated the survival of the bacteria during their transit in the GT. Interestingly, B. breve persisted until the last time point (T3), while L. salivarius did not, suggesting a species-specific survival in the GT.
Furthermore, investigating the action of the probiotic on the AD microbiota profile, we did not observe a significant change in the composition of the GM in AD children after the probiotic intake.
In conclusion, although the probiotics chosen for this study, did not shape the entire GM profile, we can confirm the ability of these species to pass the GT and to persist (only B. breve) in the GM. Moreover, our study revealed that the GM of children with AD is characterised by a dysbiotic status with a prevalence of some species such as Faecalibacterium, Oscillospira, Bacteroides, Parabacteroides and Sutterella, that can act as possible biomarkers associated to the disease. We also identified a reduction or complete absence of some microbes (i.e., Bifidobacterium, Blautia, Coprococcus, Eubacterium and Propionibacterium) with anti-inflammatory effects or involved in immune homeostasis, which might have a protective role against AD. Differences in microbiota composition between AD and CTRL could suggest to take into consideration, in future intervention studies, the species depleted or absent in AD gut microbiota as potential probiotic candidate.
Supplementary information
Acknowledgements
The authors thank the experts from BioMed Proofreading LLC for their English revision. This work was supported by the Ministry of Health, 201603X003888 assigned to LP and Ricerca Corrente 201502P003534, 201602P00370, 201503X003570 “Management of Atopic dermatitis” assigned to MEH.
Author Contributions
S.R. data acquisition, data analysis and interpretation, manuscript writing; F.D.C. healthy subject enrollment and sample collection, manuscript revising; A.Q. data analysis and manuscript revising; S.G. patient recruitment, sample collection, clinical data collection; P.V. study design and sample collection; A.R. data acquisition; A.F. manuscript revising; P.R. study conception and design; L.P. study conception and design, manuscript revising, M.E. study conception and design.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lorenza Putignani and May El Hachem jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41149-6.
References
- 1.Tan THT, Ellis JA, Saffery R, Allen KJ. The role of genetics and environment in the rise of childhood food allergy. Clin. Exp. Allergy. 2012;42:20–29. doi: 10.1111/j.1365-2222.2011.03823.x. [DOI] [PubMed] [Google Scholar]
- 2.Flohr C, Mann J. New insights into the epidemiology of childhood atopic dermatitis. Allergy. 2014;69:3–16. doi: 10.1111/all.12270. [DOI] [PubMed] [Google Scholar]
- 3.Cuello-Garcia, C. A. et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Prebiotics. World Allergy Organ. J. 9 (2016). [DOI] [PMC free article] [PubMed]
- 4.Fiocchi, A. et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 8 (2015). [DOI] [PMC free article] [PubMed]
- 5.Noverr MC, Huffnagle GB. The ‘microflora hypothesis’ of allergic diseases. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2005;35:1511–1520. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. Inducible Foxp(3+) regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vernocchi P, et al. Understanding probiotics’ role in allergic children: the clue of gut microbiota profiling. Curr. Opin. Allergy Clin. Immunol. 2015;15:495–503. doi: 10.1097/ACI.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 8.Cosmi L, Liotta F, Maggi E, Rornagnani S, Annunziato F. Th17 and Non-Classic Th1 Cells in Chronic Inflammatory Disorders: Two Sides of the Same Coin. Int. Arch. Allergy Immunol. 2014;164:171–177. doi: 10.1159/000363502. [DOI] [PubMed] [Google Scholar]
- 9.Elazab N, et al. Probiotic Administration in Early Life, Atopy, and Asthma: A Meta-analysis of Clinical Trials. Pediatrics. 2013;132:E666–E676. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 10.Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, Prebiotics and Immunomodulation of Gut Mucosal Defences: Homeostasis and Immunopathology. Nutrients. 2013;5:1869–1912. doi: 10.3390/nu5061869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Probiotics in food: health and nutritional properties and guidelines for evaluation. (Food and Agriculture Organization of the United Nations: World Health Organization, 2006).
- 12.Winkler P, Ghadimi D, Schrezenmeir J, Kraehenbuhl JP. Molecular and cellular basis of microflora-host interactions. J. Nutr. 2007;137:756S–772S. doi: 10.1093/jn/137.3.756S. [DOI] [PubMed] [Google Scholar]
- 13.Huang R, et al. Probiotics for the Treatment of Atopic Dermatitis in Children: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Front. Cell. Infect. Microbiol. 2017;7:392. doi: 10.3389/fcimb.2017.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S-O, et al. Effects of probiotics for the treatment of atopic dermatitis: a meta-analysis of randomized controlled trials. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2014;113:217–226. doi: 10.1016/j.anai.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Putignani L, et al. MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol Biosyst. 2011;7:620–9. doi: 10.1039/C0MB00138D. [DOI] [PubMed] [Google Scholar]
- 16.Ying YX, Ding WL, Li Y. Characterization of Soil Bacterial Communities in Rhizospheric and Nonrhizospheric Soil of Panax ginseng. Biochem. Genet. 2012;50:848–859. doi: 10.1007/s10528-012-9525-1. [DOI] [PubMed] [Google Scholar]
- 17.Ercolini D, De Filippis F, La Storia A, Iacono M. ‘Remake’ by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl Env. Microbiol. 2012;78:8142–5. doi: 10.1128/AEM.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. in. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. in Nat Methods. 2010;7:668–9. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 21.Rapid Cycle Real-Time PCR, 10.1007/978-3-642-59524-0 (Springer Berlin Heidelberg, 2001).
- 22.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- 25.Toh ZQ, Anzela A, Tang ML, Licciardi PV. Probiotic therapy as a novel approach for allergic disease. Front Pharmacol. 2012;3:171. doi: 10.3389/fphar.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nylund L, Satokari R, Salminen S, de Vos WM. Intestinal microbiota during early life - impact on health and disease. Proc. Nutr. Soc. 2014;73:457–469. doi: 10.1017/S0029665114000627. [DOI] [PubMed] [Google Scholar]
- 27.Abrahamsson TR, et al. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012;129:434–U244. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 28.Marrs T, Flohr C. How do Microbiota Influence the Development and Natural History of Eczema and Food Allergy? Pediatr Infect J. 2016;35:1258–1261. doi: 10.1097/INF.0000000000001314. [DOI] [PubMed] [Google Scholar]
- 29.Nylund L, et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 30.West CE, Jenmalm MC, Prescott SL. The gut microbiota and its role in the development of allergic disease: a wider perspective. Clin Exp Allergy. 2015;45:43–53. doi: 10.1111/cea.12332. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez JM, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berni Canani R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. Isme J. 2016;10:742–50. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Candela M, et al. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nylund, L. et al. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. Bmc Microbiol. 13 (2013). [DOI] [PMC free article] [PubMed]
- 35.Watanabe S, et al. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J Allergy Clin Immunol. 2003;111:587–91. doi: 10.1067/mai.2003.105. [DOI] [PubMed] [Google Scholar]
- 36.Miquel S, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–61. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137:852–60. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gophna U, Konikoff T, Nielsen HB. Oscillospira and related bacteria - From metagenomic species to metabolic features. Env. Microbiol. 2017;19:835–841. doi: 10.1111/1462-2920.13658. [DOI] [PubMed] [Google Scholar]
- 40.Konikoff T, Gophna U. Oscillospira: a Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016;24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Hua X, Goedert JJ, Pu A, Yu G, Shi J. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. EBioMedicine. 2016;3:172–179. doi: 10.1016/j.ebiom.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odamaki T, et al. Distribution of different species of the Bacteroides fragilis group in individuals with Japanese cedar pollinosis. Appl. Environ. Microbiol. 2008;74:6814–6817. doi: 10.1128/AEM.01106-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. 2002;51:51–55. doi: 10.1136/gut.51.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sipka S, Bruckner G. The immunomodulatory role of bile acids. Int Arch Allergy Immunol. 2014;165:1–8. doi: 10.1159/000366100. [DOI] [PubMed] [Google Scholar]
- 45.Curtis MM, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 49.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho GS, et al. Quantification of Slackia and Eggerthella spp. in Human Feces and Adhesion of Representatives Strains to Caco-2 Cells. Front Microbiol. 2016;7:658. doi: 10.3389/fmicb.2016.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morinaga S, Sakamoto K, Konishi K. Antitumor activity and its properties of Eubacterium lentum. Jpn. J. Cancer Res. Gann. 1988;79:117–124. doi: 10.1111/j.1349-7006.1988.tb00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. Isme J. 2014;8:1323–35. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shu M, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PloS One. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macia L, et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev. 2012;245:164–76. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 56.De Benedetto A, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127:773-86.e1–7. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parvez S, Malik KA, Ah Kang S, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–85. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 58.Vitali B, et al. Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol. 2010;10:4. doi: 10.1186/1471-2180-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue Y, Iwabuchi N, Xiao J-Z, Yaeshima T, Iwatsuki K. Suppressive effects of bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull. 2009;32:760–763. doi: 10.1248/bpb.32.760. [DOI] [PubMed] [Google Scholar]
- 60.Drago L, De Vecchi E, Gabrieli A, De Grandi R, Toscano M. Immunomodulatory Effects of Lactobacillus salivarius LS01 and Bifidobacterium breve BR03, Alone and in Combination, on Peripheral Blood Mononuclear Cells of Allergic Asthmatics. Allergy Asthma Immunol. Res. 2015;7:409–413. doi: 10.4168/aair.2015.7.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalliomaki M, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–9. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 62.Fiocchi A, et al. Clinical Use of Probiotics in Pediatric Allergy (CUPPA): A World Allergy Organization Position Paper. World Allergy Organ J. 2012;5:148–67. doi: 10.1097/WOX.0b013e3182784ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim JY, et al. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: a double-blind, randomized, placebo-controlled trial. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2010;21:e386–393. doi: 10.1111/j.1399-3038.2009.00958.x. [DOI] [PubMed] [Google Scholar]
- 64. Licari, A. et al. Atopic dermatitis: is there a role for probiotics? J. Biol. Regul. Homeost. Agents. 29, 18–24 (2015). [PubMed]
- 65. Kirjavainen, P. V., Salminen, S. J. & Isolauri, E. Probiotic bacteria in the management of atopic disease: underscoring the importance of viability. J. Pediatr. Gastroenterol. Nutr. 36, 223–227, 10.1097/00005176-200302000-00012 (2003). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.